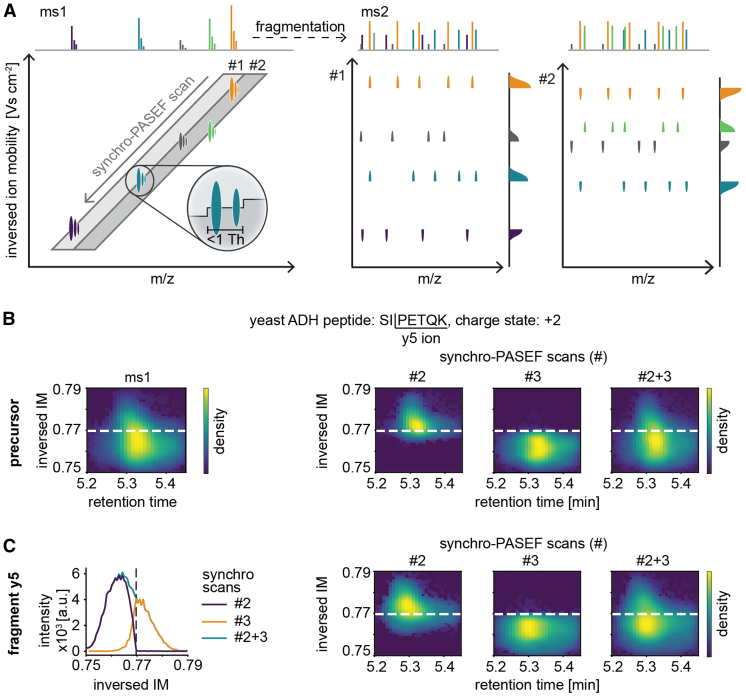
Fig. 4.
Principle of precursor slicing in synchro-PASEF.A, two synchro scans and five precursors are schematically depicted (left panel). As the trailing edge of synchro scan #1 moves through the orange precursor, the top parts of all fragments appear at the same time, followed by the bottom part in scan #2 (middle and right panels, respectively). The slicing position is known for each precursor, and all pieces with the same slicing position belong together. This specifies the precursor–fragment and fragment–fragment relationships. B, slicing of the quadrupole of the yeast ADH peptide SIPETQK. For illustration, minimal collision energy was applied. The precursor peak can be quantitatively reconstructed from the sliced parts in two subsequent synchro scans. C, same as (B) but applying collision energy and focusing on the y5 ion. The traces in adjacent synchro scans add up to the full trace for this transition (left panel). The fragment is sliced in exactly the same way as the precursor in (B) (right three panels). ADH, alcohol dehydrogenase; PASEF, parallel accumulation—serial fragmentation.