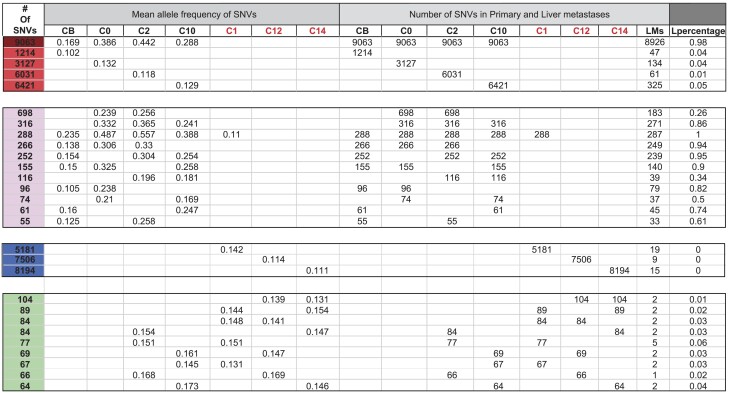
Table 1.
Grouping of mutations into clones in Case 1.
|
The single-nucleotide variations (SNVs) in the 7 whole-genome sequencing (WGS) samples of Case1 are grouped according to the clonal patterns. Each clone, representing a group of mutations, is named by the number of mutations associated with the clone. Each row represents such a clone. Clones having similar geographical patterns are further partitioned colored categories, reddish (maroon, red and pink), blue and green. Columns of the table are divided into three sections: (1) Clonal frequency in the seven WGS samples; (2) number of mutations in these seven samples; (3) number of mutations in the samples from the liver metastases; and (4) (the last two columns) the number and proportion of the mutations observed in the liver metastases (L denoting the metastases).
