Fig. 3: Nucleic acid binding and ATPase status of hexameric helicases.
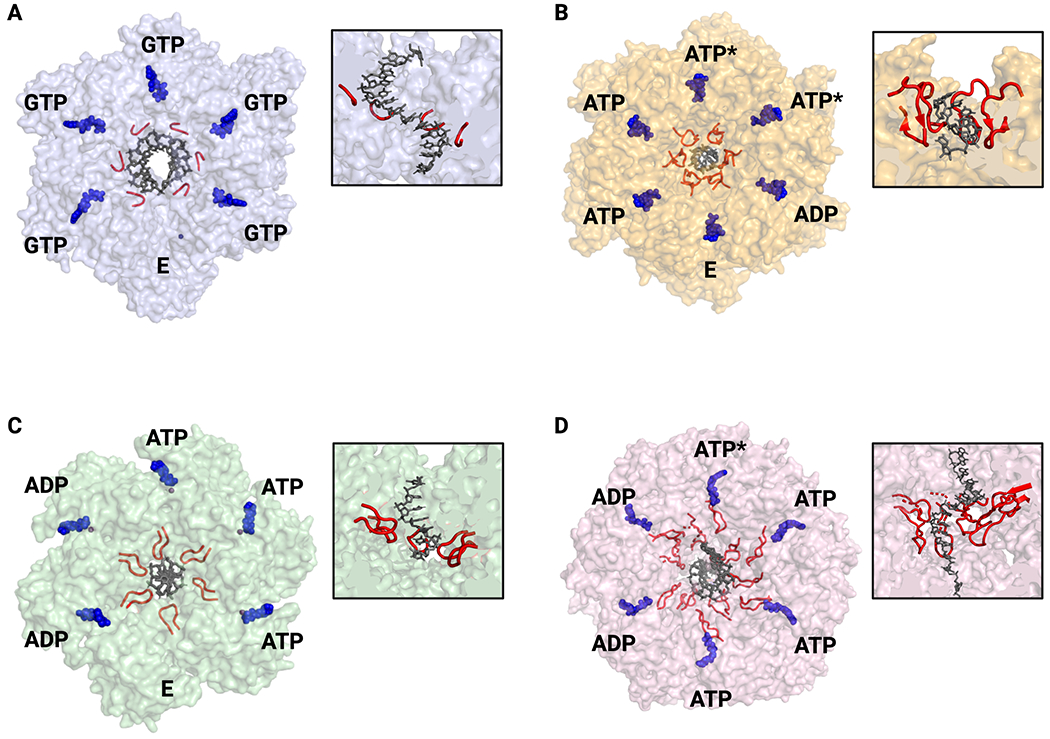
A. SF-IV – G. stearothermophilus DnaB. All GTP-bound active sites are in the same conformation and thus ATPase status around the ring cannot be assigned (PDB 4ESV). B. SF-V – E. coli Rho with ATPase state assigned by active site conformation and nucleotide status (PDB 3ICE). C. SF-III – papillomavirus E1 with ATPase state assigned by active site conformation (PDB 2GXA). D. SF-VI – D. melanogaster Mcm2-7 with ATPase state assigned by cryo-EM density and active site conformation (PDB 6RAZ). For all – nucleic acid substrates are shown in blue and pore loops in red. Nucleotide states: ATP – substrate bound; ADP – product bound; E – empty/exchangeable, ATP* - hydrolysis competent. All views are looking down from the 3’-5’ perspective. Insets show cutaway of helicase structure to highlight pore loops interactions with nucleic acid substrates.
