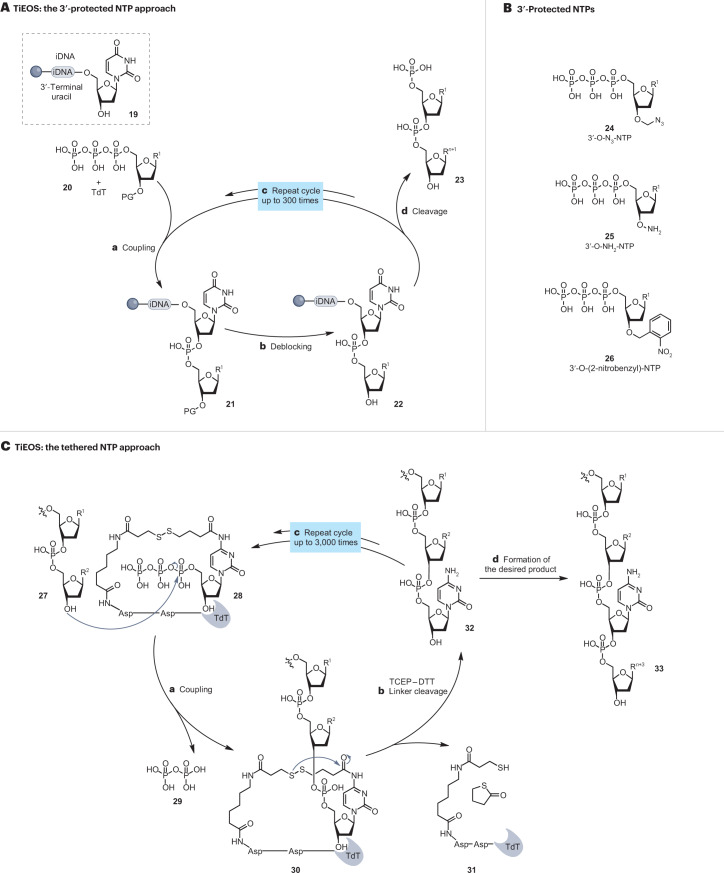
Fig. 3. Mechanisms for TiEOS.
A, Schematic representation of 3ʹ-protected nucleoside 5ʹ-triphosphate (NTP) approach. Resin beads are pre-loaded with an initiator DNA (iDNA) 19 to provide a template for binding of terminal deoxynucleotidyl transferase (TdT) and as a post-synthesis cleavage site27,72,77,78,95. Oligonucleotide synthesis then proceeds in a stepwise fashion in the 5ʹ-to-3ʹ direction. TdT ligates NTP 20 to the 3ʹ terminus of the growing oligonucleotide chain with each NTP protected at 3ʹ-OH with a protecting group (PG) 24–26 (refs. 27,28,82). The resin is washed to remove surplus reagents and the pyrophosphate by-product of the ligation. After deblocking or deprotection of the 3ʹ-PG (step b), the resin-bound 3ʹ-OH nucleophile of 22 becomes available for the next synthesis cycle (step c). The complete sequence is assembled by repeating the cycle of TdT-catalysed NTP(PG) coupling (step a) and deblocking (step b). On completion, the synthesized oligonucleotide 4 is cleaved from the solid support (step d) by uracil DNA glycosylase. B, Examples of NTP(PG)s used in the method — 3ʹ-azidomethyl-protected NTPs 24 by Nuclera Nucleics, Molecular Assemblies, 3ʹ-ONH2-protected NTPs 25 by DNA Script and 3ʹ-O-2-nitrobenzyl 26 by Camena Bioscience97,100,101,108,118. C, Schematic representation for alternative (tethered) protecting strategies. 3ʹ-Unprotected NTPs (cytidine) are supplied pre-immobilized within the TdT-active site 28, via a short and labile linker121,129. TdT then catalyses the incorporation of this NTP into the growing DNA strand 30 (step a) and sterically prevents the uncontrolled polymerization of the NTP until the linker is cleaved (step b), releasing the oligonucleotide 32. The cycle is repeated (step c) until the desired oligonucleotide 33 is completed (step d). Asp, aspartic acid; DTT, dithiothreitol; TCEP, tris-carboxyethylphosphine; TiEOS, template-independent enzymatic oligonucleotide synthesis.