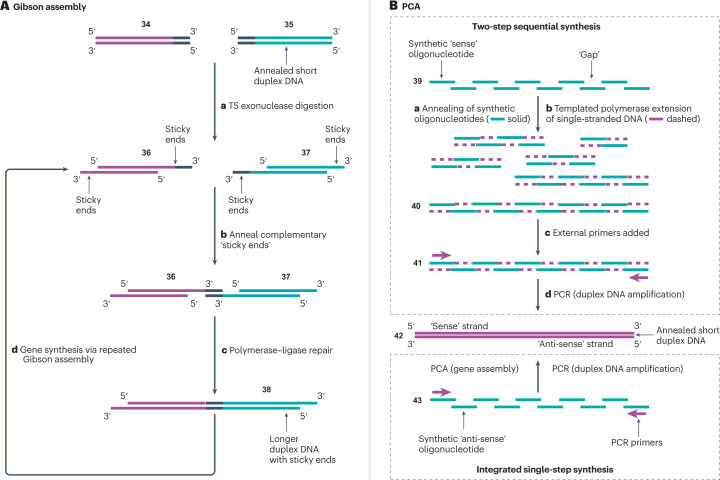
Fig. 4. Schematic representations of assembly methods for DNA synthesis.
A, Gibson assembly25,139,140. Two duplex DNA strands 34 and 35 are selected with a complementary terminal overlap region (black). Digestion with T5 exonuclease (step a) degrades each strand of the DNA duplexes in the 5ʹ-to-3ʹ direction, yielding the sticky ends of 36 and 37, followed by the annealing of complementary sticky ends between the two DNA duplexes143 (step b). Phusion polymerase and Taq ligase are then combined (step c) to ligate the two short DNA duplexes into a single, long DNA duplex construct 38 (refs. 144,145). The process is repeated (step d) for gene assembly. B, Polymerase cycling assembly (PCA)26,141. High-purity synthetic oligonucleotides 39 are designed, such that annealing of complementary overlaps generates the desired long duplex DNA construct (step a). The desired construct is then assembled in either a single step from 43 or two steps from 39 using a DNA polymerase (step b) to yield template 40. PCR amplification (steps c and d) of 41 amplifies the desired long duplex DNA construct 42.