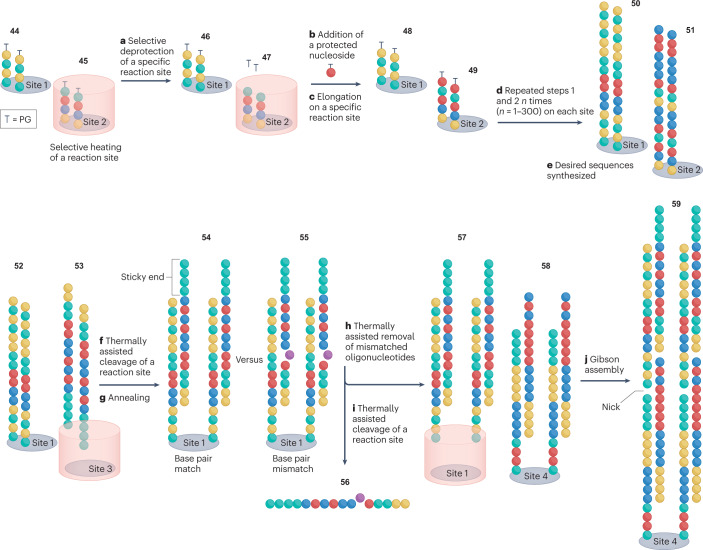
Fig. 5. Schematic representation of thermally controlled DNA synthesis.
Terminally protected oligonucleotide strands 44 and 45 are immobilized on discrete reaction sites (sites 1 and 2). Thermal heating of a chosen site (site 2) cleaves terminal protecting groups (PGs) 47 (step a), enabling selective elongation of strands on this site29,51,52,151 (steps b and c) Elongation of a desired oligonucleotide is performed via TiEOS or the phosphoramidite method to selectively generate 49 (refs. 27,37). Repeated steps a–c on other selected reaction sites (for example, site 1) sequentially produce bespoke oligonucleotides 50 (steps d and e). Thermally assisted reagent treatment of selected sites cleaves safety catch linkers and liberates oligonucleotides 53 from a chosen site29,151 (step f). The liberated oligonucleotides (step g) anneal to complementary chip-bound oligonucleotides 52 producing perfectly annealed double-stranded DNA 54, which has a higher denaturation temperature than that of DNA duplexes formed by oligonucleotides with mismatches 55. The heating of reaction site 1 allows the mismatched oligonucleotides 56 to be washed away150 (step h). Thermally assisted reagent treatment of site 1 cleaves safety catch linkers and releases the desired duplex DNA 57 from the chip29,151 (step i). This liberated DNA is annealed to a chip-bound complementary DNA duplex 58 to form the nicked construct 59 (step j). The process is repeated to elongate the double-stranded DNA until the desired gene is assembled.