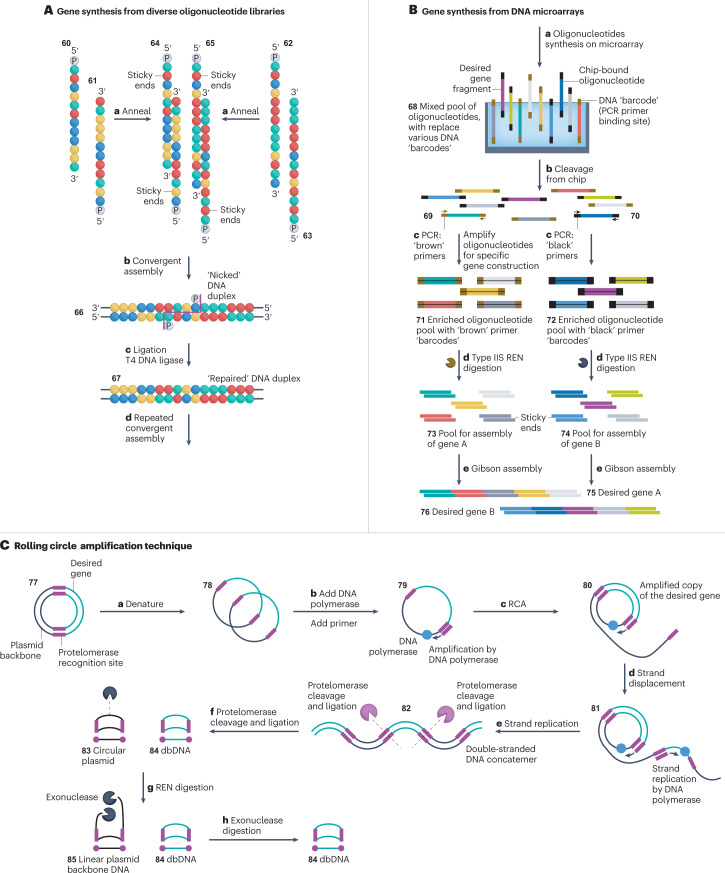
Fig. 6. Gene synthesis strategies.
A, Gene synthesis from diverse oligonucleotide libraries. 5ʹ-Phosphorylated sense oligonucleotide strands 60 and 61 are annealed (step a) to complementary 5ʹ-phosphorylated antisense strands 62 and 63. The resulting DNA duplexes 64 and 65 have two 5ʹ-overhangs or sticky ends that are used to anneal (step b) the duplexes into an extended, ‘nicked’ duplex 66 (‘nick’ highlighted in magenta). T4 DNA ligase is then used (step c) to stitch the oligonucleotides at the nick site into an elongated, larger DNA duplex 67. Cycles of annealing and ligation are repeated until the desired gene is assembled153–155 (step d). B, DNA microarrays30. A library of bespoke single-stranded oligonucleotides 68 is generated with 3ʹ-terminal and 5ʹ-terminal DNA ‘barcodes’ on a miniaturized chip30,47,48,50,164,166,167 (step a). These sequences are cleaved (step b) from the microchip to yield a pool of template oligonucleotides with a range of DNA ‘barcodes’ (only two, black and brown, ‘barcodes’ are shown for clarity)164,167. Primers selectively anneal to either the ‘brown’ 69 or ‘black’ 70 DNA barcodes and specifically amplify oligonucleotides via PCR (step c), according to the identity of the barcode at its 3ʹ and 5ʹ termini146–148. The resulting duplex DNA constructs 71 and 72 still contain the DNA barcodes at their termini, which must be removed prior to gene assembly. DNA barcodes are cleaved (step d) from the duplex DNA 71 and 72 by type IIS restriction endonucleases (REN), giving rise to assembly pools of sequences 73 and 74 with sticky ends30. Duplex DNA fragments are annealed (step e) via complementary sticky ends and assembled into desired genes 75 and 76 via Gibson assembly25,139,140. C, Rolling circle amplification (RCA)175. Template plasmid DNA 77 with a desired gene cassette (green) and protelomerase sites (magenta) is thermally denatured (step a) to create a single-stranded template 78 (ref. 178). A complementary primer binds to the protelomerase sites 79 (step b) and the template is amplified via RCA (steps c–e), to produce double-stranded concatemeric DNA 82 with alternating copies of the desired cassette (green) and the unwanted plasmid backbone (black)176,181–183. Protelomerase then cuts (step f) the duplex at its recognition sites and ligates the cut ends generating covalently closed ‘doggybone’ DNA (dbDNA) 84 and a circular plasmid DNA 83 as a by-product. The circular backbone of the plasmid DNA is subsequently cut (step g) by REN and digested (step h) by exonucleases179.