Fig. 2: APOE4 alters cholesterol homeostasis and localization in human post-mortem oligodendrocytes.
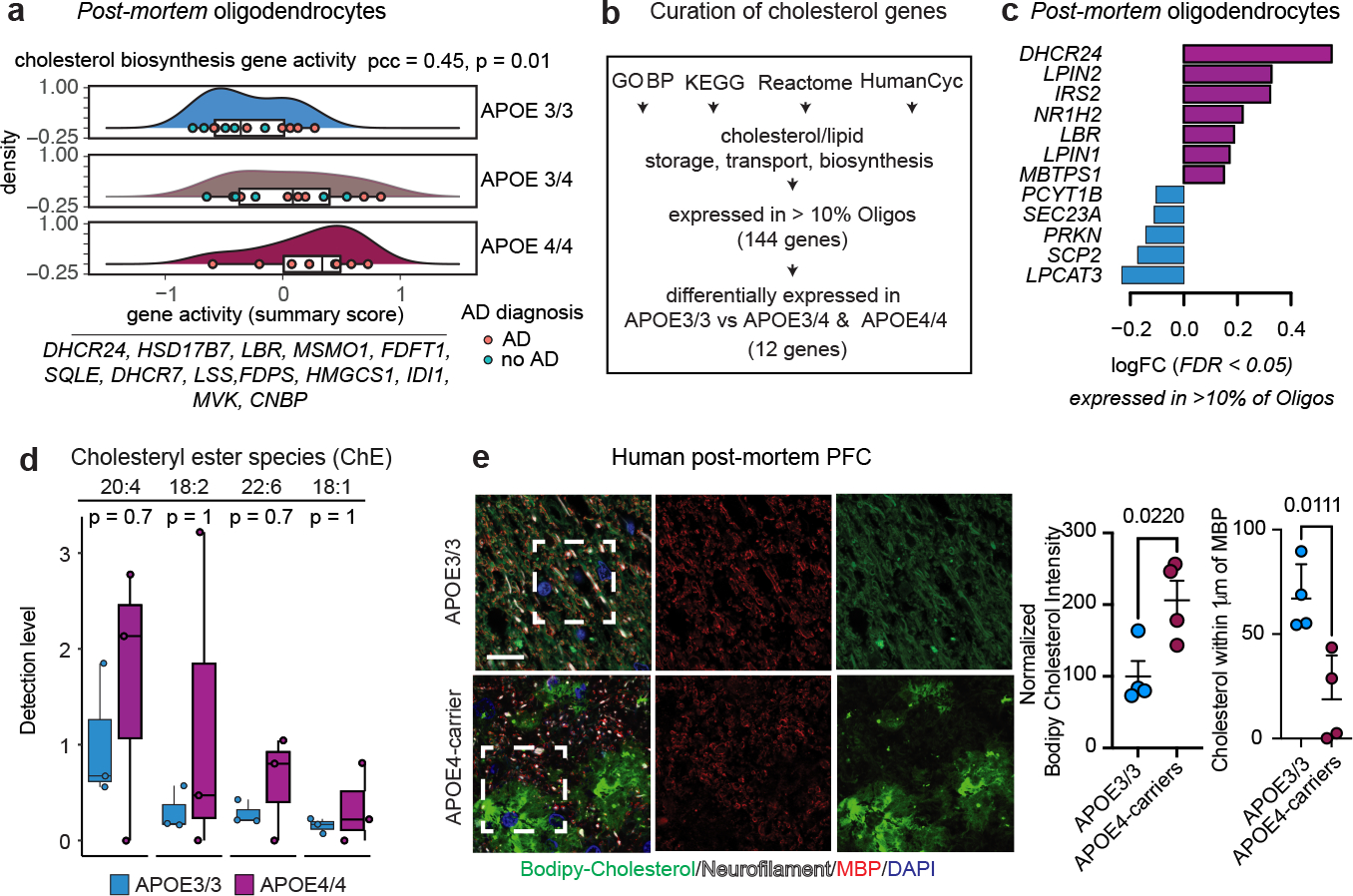
a, Dose-dependent association between APOE4 and cholesterol biosynthesis genes aggregated expression in oligodendrocytes (APOE 4/4 > 3/4 > 3/3, Pearson correlation coefficient (pcc) = 0.45, p-value = 0.01, two-sided). b, Curation process of cholesterol-related genes. Pathway databases were filtered using the terms cholesterol/lipid storage, transport, or synthesis. c, Cholesterol-related genes differentially expressed in APOE3/3 vs APOE3/4 and APOE4/4 post-mortem oligodendrocytes (snRNA-seq data, FDR-adjusted p-value < 0.05, negative binomial mixed model). d, Detection levels of four cholesteryl esters quantified by mass-spectrometry of post-mortem human corpus callosum from APOE3 (APOE3/3, females, n=3) and APOE4-carriers (APOE4/4, females, n=3). Data points represent relative abundance per individual (n=3 individuals per genotype). Numbers in the top label cholesteryl ester species. First number specifies the carboxylate position connecting the fatty acid to the cholesterol hydroxyl group. Second number specifies the frequency of unsaturated fatty acid bonds. Two-sided unadjusted wilcoxon test p-values are shown. Boxplots indicate median, 25th and 75th percentiles. e, Representative images of Bodipy-cholesterol staining and anti-myelin basic protein (MBP) immunoreactivity in prefrontal cortex (BA10) from APOE4-carriers compared to APOE3/3 individuals. Scale bar 10 μm. Cholesterol localization was analyzed across 4 individuals for each APOE genotype. Right panels show total Bodipy-cholesterol signal and percent of Bodipy-cholesterol signal within 1 μm of an MBP-positive axon quantified using Imaris software. Data points represent individuals. Bars depict means, error bars represent standard error of the mean, and p values were calculated with an unpaired two-tailed student’s t-test.
