Extended Data Fig. 1. Subject-level metadata and single-cell annotation quality control.
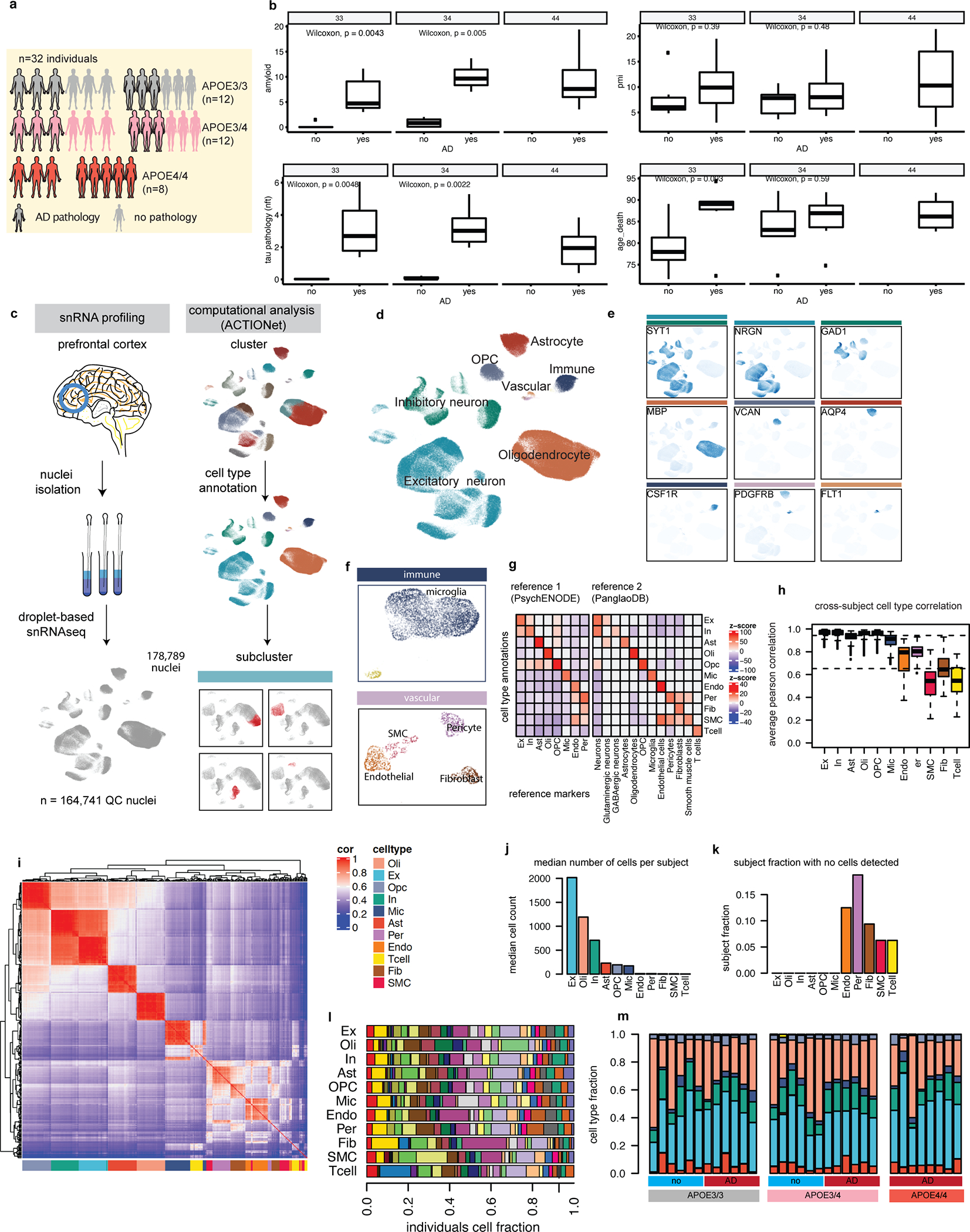
a, Study cohort description by APOE genotype groups: APOE3/3-carriers (gray), APOE3/4-carriers (pink), APOE4/4-carriers (red); balanced according to positive (glow) or negative pathological diagnosis (no glow) Cartoons generated with BioRender. APOE4/4 group comprised 3 male and 5 female subjects, all with an AD diagnosis. AD status is defined based on pathological (high amyloid and tau pathology) and cognitive diagnosis of Alzheimer’s dementia (final consensus cognitive diagnosis, cogdx=4). b, Distribution of pathology variables, PMI, and age at death in cohort. Unadjusted wilcoxon test p-values shown, two-sided (N = 6 per group, except for APOE4/4, where N = 8). Boxplots indicate median, 25th and 75th percentiles. c, Experimental and computational workflow of the single-cell analysis involves single nuclei isolation and sequencing, followed by computational analysis for sub-clustering and cell type annotation. Cartoons generated with BioRender d, Two-dimensional representation of all high-quality cells included in downstream analysis labeled by major cell types. e, Cell-type-specific marker gene expression projected onto two-dimensional representation of cell space. Colored bars indicate the cell type for which a given gene was considered a marker. See panel i for legend. f, Expanded view of immune and vascular cell types within cell space. g, Enrichment of markers from two independent datasets (columns) within genes with preferential gene expression across annotated cell groups (rows). h, Distribution of inter-subject correlation values by cell type. Boxplots indicate median, 25th and 75th percentiles. i, Pairwise correlations of cell-type-specific individual-level transcriptomic profiles (average expression values across cells of a given type, N = 32 profiles per cell type). j, Median number of cells per subject by cell type. k, Fraction of subjects lacking cells of a given type. l, Individual distributions across cell types. m, Individual cell-type fractions organized by pathological diagnosis and APOE genotype.
