Extended Data Fig. 2: APOE-associated and lipid pathway changes in APOE4 and AD.
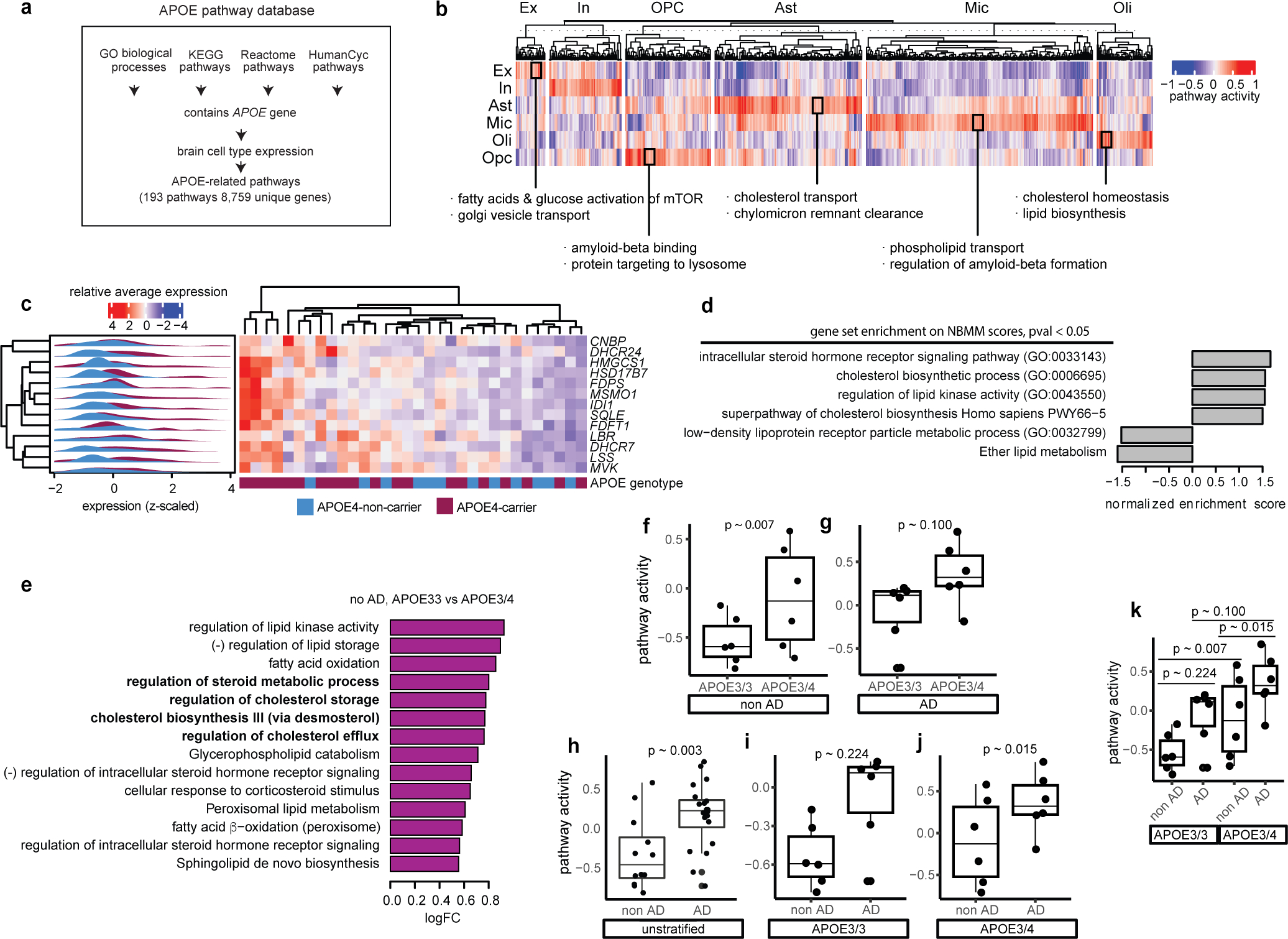
a, Curation process for APOE-associated pathway database. Brain cell type expression was defined as nonzero detection of a gene in >10% of cells of that cell type. b, Transcriptional activity scores of APOE-associated pathways that show cell-type-specific patterns. c, Individual-level average expression of cholesterol biosynthesis genes from Fig. 2a in oligodendrocytes. d, Overrepresentation of lipid-related pathways (database from Fig. 1c) within genes differentially expressed in APOE4 relative to APOE3 in human post-mortem oligodendrocytes as estimated by a negative binomial mixed model (NBMM). e, Altered lipid-associated pathways (see methods) in APOE3/3 vs APOE3/4-carriers, in no AD background (control) (nominal p-value < 0.05, linear model, unadjusted, N = 6 per group). Pathways of special interest are highlighted in bold. (−) indicates negative regulation. f-k, Pathway activity scores for ‘Cholesterol biosynthesis III (via desmosterol) stratified by APOE genotype and/or AD pathology (p-values, linear model, unadjusted, n = 6 per group). Boxplots indicate median, 25th and 75th percentiles.
