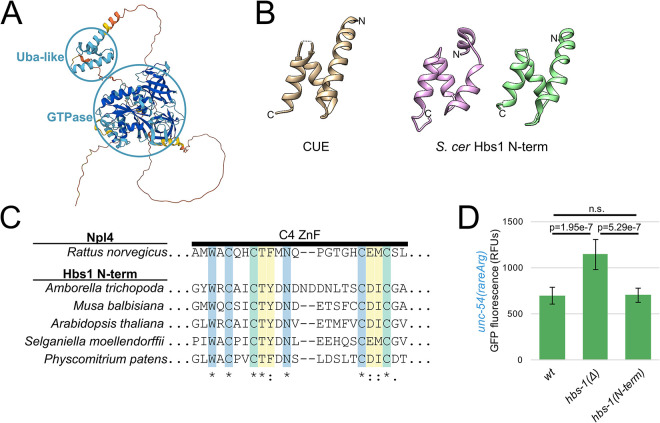
Fig 6. HBS-1 N-terminus resembles a ubiquitin-binding domain and is dispensable for NGD.
(A) H. sapiens Hbs1 protein structure as predicted by AlphaFold [29]. Prediction confidence is as in Fig 5A, with dark blue showing regions of high confidence. Confident domains are circled in blue and labeled. (B) Structural homology between Hbs1 N-terminal domain and ubiquitin-binding domains. At left is a structure representative of the UBA clan: CUE from [38] (tan, S. cerevisiae Vps9p, 1P3Q). At right are two structures of the Hbs1 N-terminus from [12] (pink, S. cerevisiae, 3IZQ) and [14] (green, S. cerevisiae, 5M1J). Amino and carboxy termini indicated with N and C, respectively. Note overall similarity in topology and fold across structures. (C) The N-terminal zinc finger (ZnF) of plant Hbs1 is homologous to the Ub-binding ZnF of rat Npl4. Multiple sequence alignment of the ZnF of Hbs1 from phylogenetically diverse plants. Residues that are highly conserved among Npl4 homologs are in blue, residues that contact Ub are in yellow, and residues that are both conserved and contact Ub are in green. Coloring and annotation of Npl4 residues from [39]. (D) Mean RFUs (relative fluorescence units) of indicated strains (n≥15 animals/strain) in the unc-54(rareArg) background. One standard deviation shown as error bars. p values from Welch’s t-test.