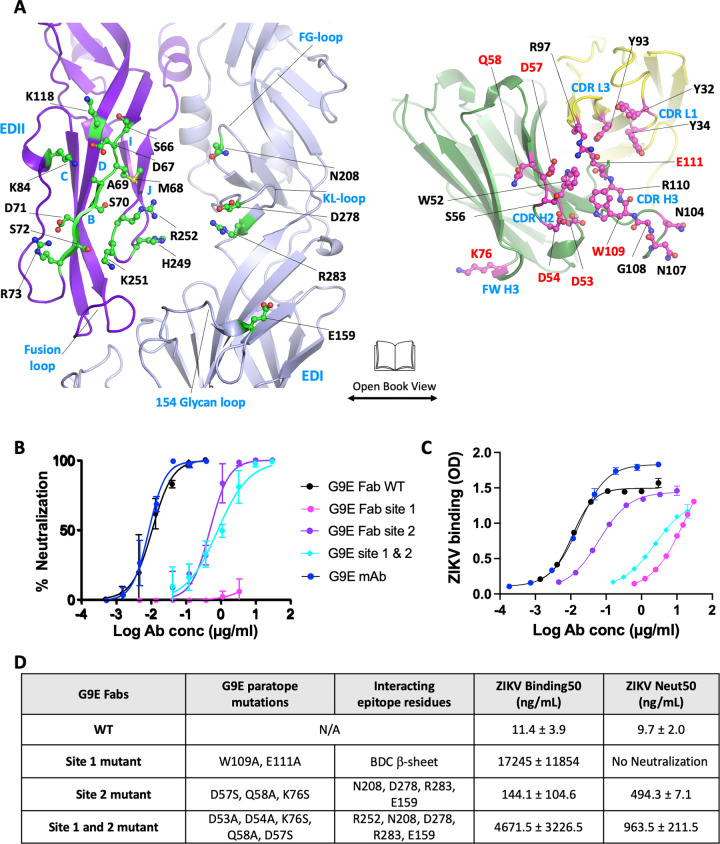
Fig 5. G9E cross-dimer binding underlies potent neutralization of ZIKV.
(A) Close up view of the interactions between ZIKV E-dimer and G9E is shown in an "open book" representation. The interfacing residues between ZIKV E-dimer and G9E were identified by Protein Interfaces, Surfaces, and Assemblies program (PISA). Contact residues on E protein (green) and G9E (pink) are shown in ball and stick representations. The paratope residues selected for site-directed mutations are highlighted in red text. (B) Representative ZIKV neutralization curves for G9E WT and paratope mutant Fabs, G9E-S1, G9E-S2, or G9E-S1/2. (C) Representative ZIKV binding curves for G9E WT and paratope mutant Fabs, G9E-S1, or G9E-S2, or G9E-S1/2. Error bars (in panel B and C) are standard deviations of technical replicates. (D) Characteristics of ZIKV binding and ZIKV neutralization by G9E WT and paratope mutant Fabs. The neutralizing and binding data (in panel D) are the means and standard deviations of EC50 values obtained from three (binding) or two (neutralizing) independent experiments with technical replicates for each titration point.