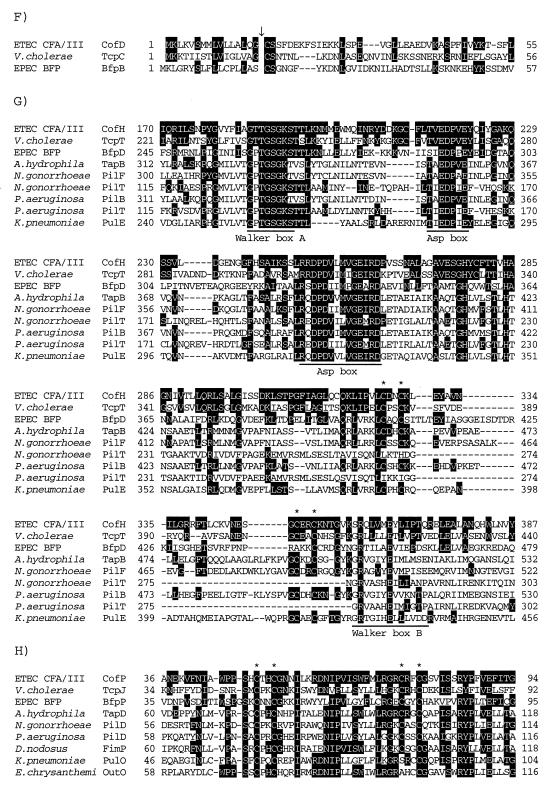
FIG. 3.
Partial alignment of the deduced amino acid sequences of the Cof proteins with those of known proteins. Identical amino acids are indicated by a black background. Gaps introduced for alignment are represented by dashes. (A) Alignment of the deduced amino acid sequences of CofR, Salmonella serovar Typhimurium PefB (GenBank accession no. L08613), ETEC FaeB (Z11709), E. coli AfaA (X76688), and E. coli PapB (X03391). (B) Alignment of the deduced amino acid sequences of CofS, ETEC FapR (X53494), ETEC CfaD (M55609), ETEC Rns (J04166), V. cholerae ToxT (X64098), EPEC PerA (Z48561), enteroaggregative E. coli (EAggEC) AggR (Z32523), S. dysenteriae VirF (X58464), and serovar Typhimurium SirC (AF134856) in the region containing the DNA-binding domain (underline). (C) Alignment of the deduced amino acid sequences of CofT, E. coli Gene X (X07264), Salmonella serovar Typhi IagB (X80892), S. flexneri IpgF (L04309), EPEC BfpH (Z68186), and E. coli Slt (M69185). Three motifs conserved in putative lytic transglycosylases are underlined. (D) Alignment of the deduced amino acid sequences of CofA, ETEC LngA (AF004308), V. cholerae TcpA (X64098), EPEC BfpA (Z68186), A. hydrophila TapA (U20255), N. gonorrhoeae PilE (X66144), P. aeruginosa PilA (M14849), Dichelobacter nodosus FimA (X52405), and Moraxella bovis TfpQ (M59712) in the N-terminal region. The cleavage site of type IV pilins is shown by a downward arrow. The conserved glycine, leucine, and glutamic acid residues are marked by asterisks. (E) Alignment of the deduced amino acid sequences of CofB, ETEC LngB, and V. cholerae TcpB (X64098) in the N-terminal region. The type IV pilin-like cleavage site is shown by a downward arrow. The conserved glycine, leucine, and glutamic acid residues are marked by asterisks. (F) Alignment of the deduced amino acid sequences of CofD, V. cholerae TcpC (X64098), and EPEC BfpB (Z68186) in the N-terminal region. The lipoprotein-cleavage site is shown by a downward arrow. (G) Alignment of the deduced amino acid sequences of CofH, V. cholerae TcpT (X64098), EPEC BfpD (Z68186), A. hydrophila TapB (U20255), N. gonorrhoeae PilF (U32588), N. gonorrhoeae PilT (S72391), P. aeruginosa PilB (M32066), P. aeruginosa PilT (M55524), and K. pneumoniae PulE (M32613) in the region containing the nucleotide-binding domain. The Walker box A, Asp boxes, and Walker box B are underlined. The conserved CXXC motifs are marked by asterisks. (H) Alignment of the deduced amino acid sequences of CofP, V. cholerae TcpJ (M74708), EPEC BfpP (Z68186), A. hydrophila TapD (U20255), N. gonorrhoeae PilD (U32588), P. aeruginosa PilD (M32066), D. nodosus FimP (U17138), K. pneumoniae PulO (M32613), and Erwinia chrysanthemi OutO (L02214) in the region containing the conserved CXXC motifs (asterisks).