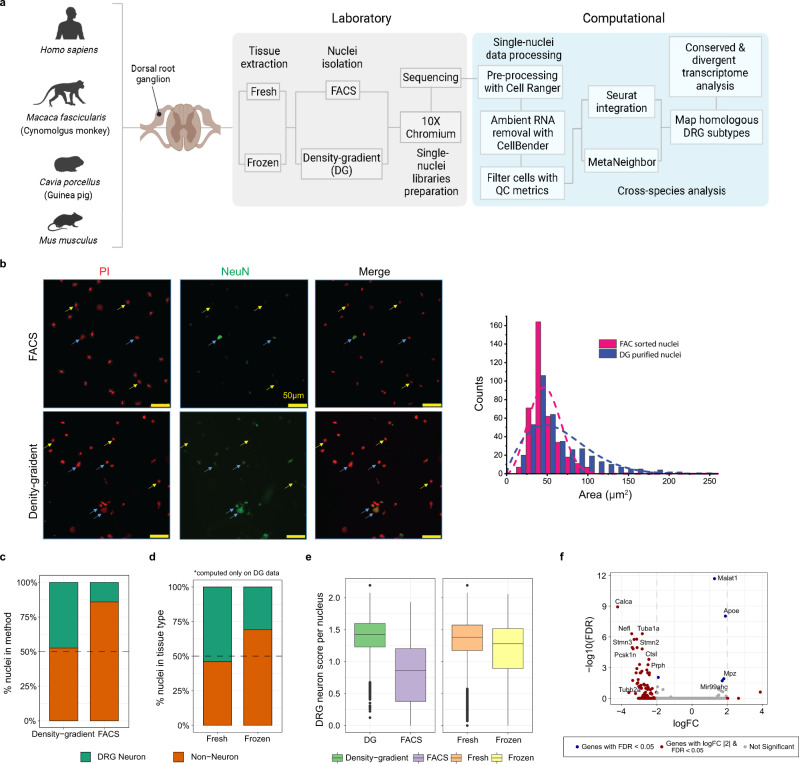
Fig. 1. Comparison of nuclei isolation methods and tissue types in dorsal root ganglia single-nucleus RNA-seq.
a Overview of laboratory and computational workflow. The graphic was created with BioRender.com b Representative micrographs of nuclei from FACS and density-gradient (DG) protocols. Blue arrows indicate neuronal nuclei (PI+ and NeuN+) and yellow arrows indicate non-neuronal nuclei (PI+ and NeuN−). Distribution of nuclei size by capture method. Dotted lines represent smoothed distributions of binned data. The experiment was repeated 4 times independently with similar results. c–e Summary metrics computed for different nuclei isolation methods and tissue types in mouse data. c, d Relative cell-type composition for the nuclei isolation methods or tissue types. Black dashed line was drawn at 50%. e DRG neuron signature score for different nuclei isolation methods and tissue types. n = 37,384 nuclei examined over 10 independent experiments. The lower and upper hinges of the boxes correspond to the 25th–75th percentile with the line in the middle depicting the median. The whiskers are set at the minimum and maximum values of the dataset. f Volcano plot showing results from differential expression analysis of transcripts from FACS versus DG nuclei. Statistically significant genes are indicated by red or blue colors. Gray dotted lines represent log-fold change thresholds of 2 and −2.