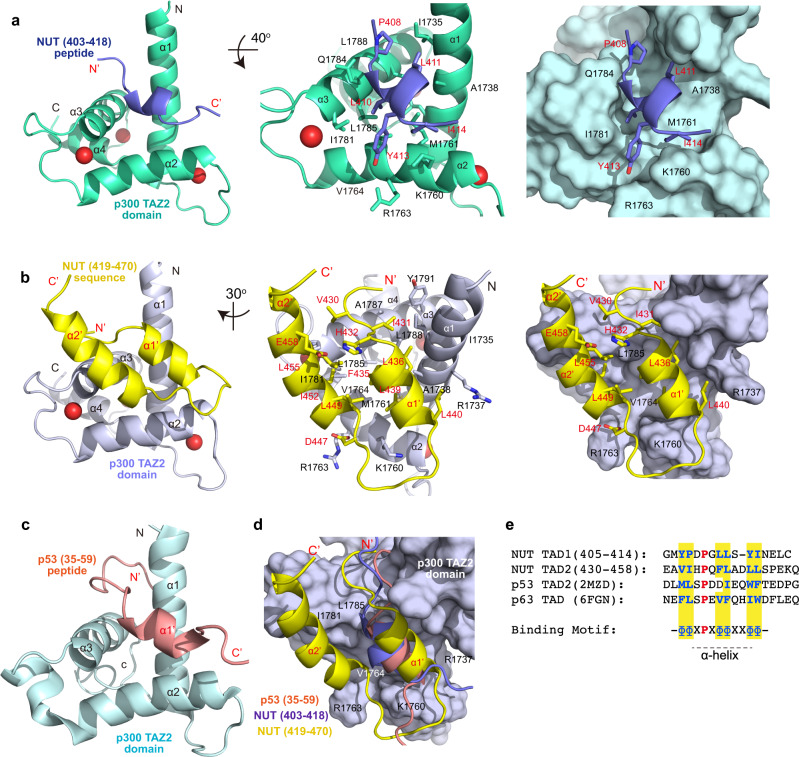
Fig. 2. Structures of p300 TAZ2 domain in complex with NUT TAD1 or TAD2.
a Interaction between p300 TAZ2 domain and F1c peptide (TAD1, residues 403–418). Left: Ribbon depiction of the lowest energy structure of the complex; Middle: Expanded diagram of key inter-molecular interactions from the α-helix F1c peptide to the hydrophobic binding core of the TAZ2 domain surrounded by α1, α2, and α3 helices. TAZ2 domain residues involved in peptide binding are labeled and colored in light-green, the peptide elements are colored in blue; Right: Surface representation of the TAZ2 domain depicting its recognition to the F1c peptide in the same orientation as (Middle). b Structure of TAZ2-NUT fusion protein containing the NUT sequence (TAD2, residues 419–470). Left: Ribbon depiction of the lowest energy structure of the fusion protein (TAZ2 in light-blue and NUT sequence in yellow); Middle: The binding core depiction of residue side-chains of TAZ2 in α1, α2, and α3 helices for hydrophobic and electrostatic interactions to the residues in the F1c sequence. Right: Surface representation of interaction positions of TAZ2-NUT fusion in the same orientation as (Middle); c The ribbon diagram of CBP TAZ2/p53 TAD2 complex. d Superposition of NUT TAD2 sequence (residues 419–470, in yellow) in the TAZ2-NUT fusion protein with NUT TAD1 sequence (residues 403–418, in blue), and p53 TAD2 (residues 35–59, in orange, 2MZD) peptides. e Sequence alignment of NUT TAD1 (residues 405–414), NUT TAD2 (residues 430–458), p53 TAD2, and p63 TAD. The conserved proline amino acid is colored in red, hydrophobic residues in blue. The binding motif common to these sequences is diagramed as, Φ represents a hydrophobic amino acid and X represents any amino acid.