Figure 8. Comparative gene regulatory networks of sea urchins and sea stars.
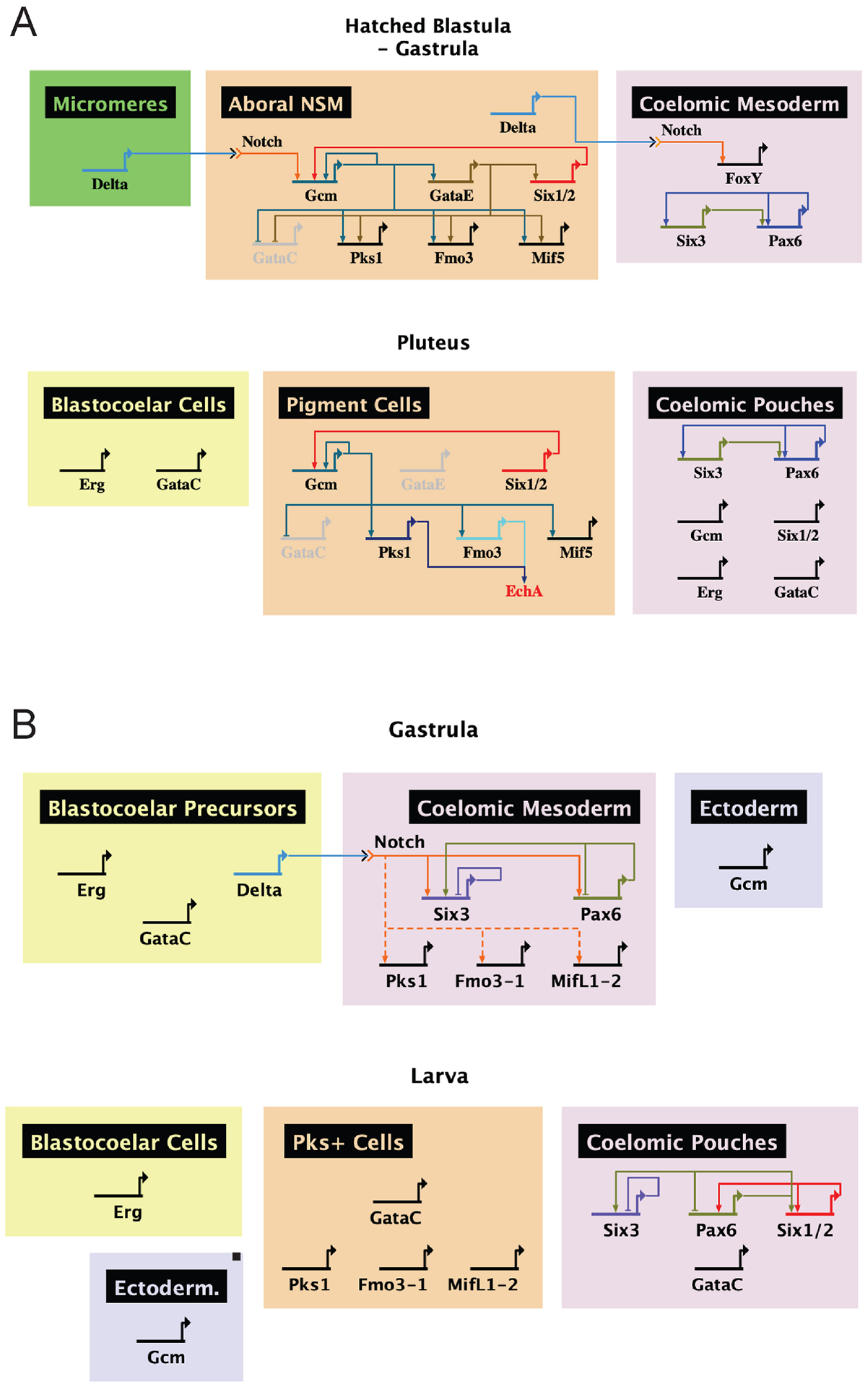
Networks were constructed using BioTapestry software. A. The sea urchin has a stable lineage of pigment cells throughout development specified by Notch. These cells are marked by SpGcm, which activates the expression of SpPks1, SpFmo3 and SpMif5. SpPks1 and SpFmo3 synthesize the pigment Echinochrome A. Relationships between genes are described elsewhere [14],[17],[18],[45],[56],[58],[62],[72]. B. The sea star lacks a lineage of cells expressing PmPks1, PmFmo3-1 and PmMifL1-2. The mesodermal expression of these genes during gastrulation is conserved, as is the requirement of Notch, though they do not depend on PmGcm for expression. Additional work is required to determine if Notch activation is direct or proceeds through an intermediate as it does in sea urchins. Later in development, PmPks1, PmFmo3-1 and PmMifL1-2 are expressed in the ectoderm with PmGataC. Relationships between genes are derived from this work and elsewhere [52].
