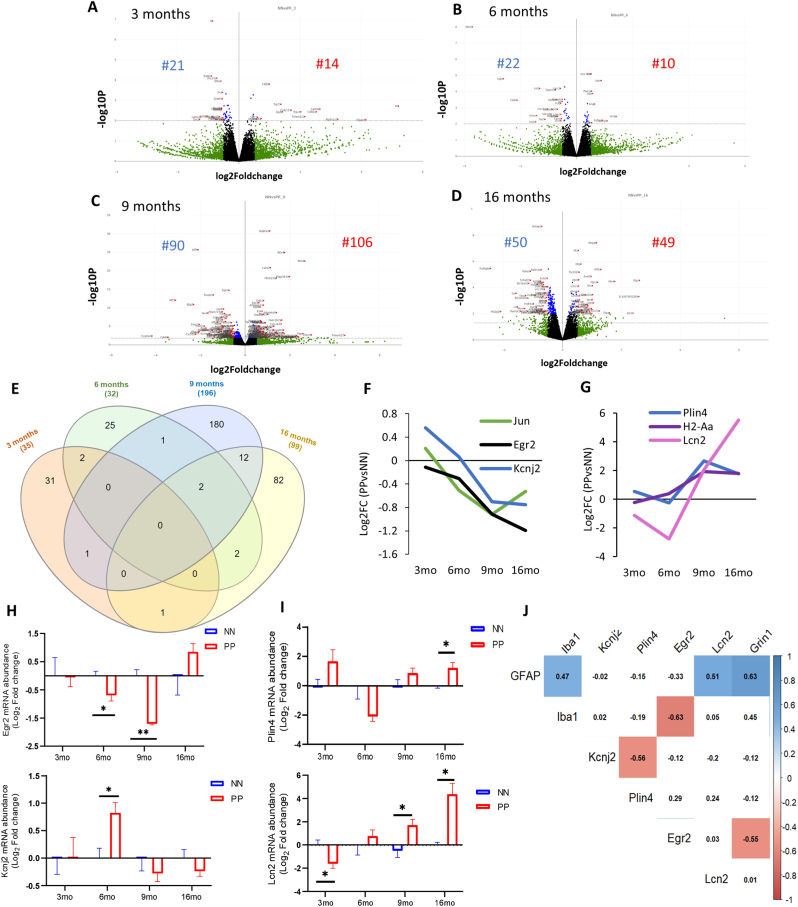
Fig. 5.
Bulk transcriptomics analysis of hippocampus in the aging PolyI:C. Gene expression changes after the prenatal and early postnatal immune challenge. Volcano plot with log2-fold change (X-axis) and -log10 p-value (Y-axis) at different age groups (A) 3 months (B) 6months (C) 9 months and (D) 16 months in PP versus NN controls. Genes with log2 fold changes >0.5 are shown in green. Significantly regulated genes are shown in red (log2 fold change >0.5, p < 0.05, left downregulated and right upregulated). Genes with insignificant log2 fold changes <0.5 are shown in black [controls (NN) n = 3/age, PolyI:C (PP) n = 3/age. (E) Venn diagram indicating the number of uniquely and commonly affected genes in the aging hippocampus following prenatal and early postnatal PolyI:C (PP) treatment. Few common genes along with their Log2 Fold changes either (F) downregulated or (G) upregulated were plotted across staging. Transcript validation analysis for (H)Egr2 and Kcnj2 and (I)Plin4 and Lcn2, *p<0.05, **p<0.005, two-way ANOVA with Benjamini Krieger and Yekutieli post hoc analyses, [controls (NN), n = 5/age, PolyI:C (PP) n = 4–5/age] (J) Spearman correlation matrix for mRNA levels (Log2FC) with highlighted significant association from 9 to 16 months among the differentially expressed cell-type markers (GFAP, Iba1 and Grin1) and Egr2, Kcnj2, Plin4 and Lcn2 between PolyI:C and saline.