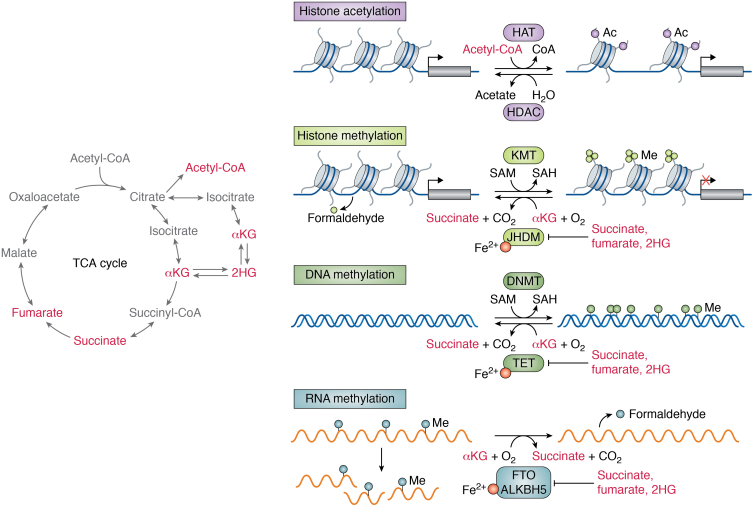
Figure 6.
Regulation of chromatin by tricarboxylic acid (TCA) cycle–associated metabolites. Certain TCA cycle–derived metabolites (left) control the regulation of chromatin and, in turn, gene expression. Histone acetyltransferases (HAT) transfer acetyl groups from acetyl-CoA to histones, thereby altering chromatin accessibility. Histone deacetylases (HDACs) remove acetyl groups from histones and generate acetate as a product. Methylation on histones and DNA is deposited by histone lysine methyltransferases (KMTs) and DNA methyltransferases (DNMTs), respectively. DNMT and KMT add methyl groups by catalyzing the transfer of a methyl group from S-adenosyl methionine (SAM), producing S-adenosyl homocysteine (SAH). Alpha-ketoglutarate (αKG)-dependent dioxygenases regulate the demethylation of histones and nucleic acids. Jumonji C-domain–containing histone demethylases (JHDMs) and ten–eleven translocation (TET) DNA methylcytosine dioxygenases, which remove repressive histone marks and 5-methylcytosine, respectively, require αKG as an obligate cosubstrate and are competitively inhibited by succinate, fumarate, and 2-hydroxyglutarate (2HG). mRNA methylation is controlled by the RNA methyltransferases and αKG-dependent dioxygenases FTO and ALKBH5, which like all αKG-dependent dioxygenases are stimulated by αKG and repressed by succinate, fumarate, and 2HG. Histone and RNA demethylation produce formaldehyde as a byproduct.