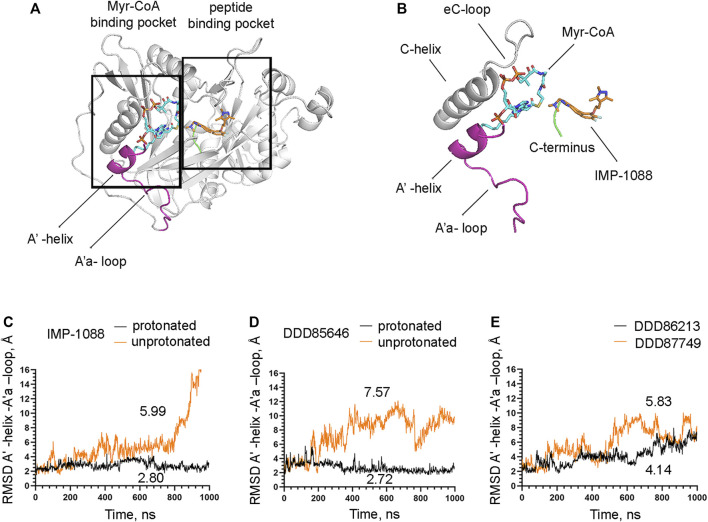
FIGURE 9.
The salt bridge stabilizes the conformation of the Myr-CoA binding pocket. (A) The crystal structure of Homo sapiens NMT1 (PDB 5MU6), depicting the Myr-CoA and peptide binding pockets (B) A zoomed view on the image on the left showing details of the Myr-CoA binding pocket. The A′-helix and A’a-loop, shown in magenta, take part in the formation of the bottom of the pocket (C) RMSD in angstroms (Å) of the heavy atoms of the A′-helix and A’a-loop vs. time of MD simulations in nanoseconds (ns) of its complexes with the protonated and unprotonated forms of IMP-1088. (D) RMSD in angstroms (Å) of the heavy atoms of the A′-helix and A’a-loop vs. time of MD simulations in nanoseconds (ns) of its complexes with the protonated and unprotonated forms of DDD85646. (E) RMSD in angstroms (Å) of the heavy atoms of the A′-helix and A’a-loop vs. time of MD simulations in nanoseconds (ns) of its complexes with DDD86213 (protonated form) and DDD87749. The numbers in the plots indicate the average RMSD in angstroms (Å).