Figure 5.
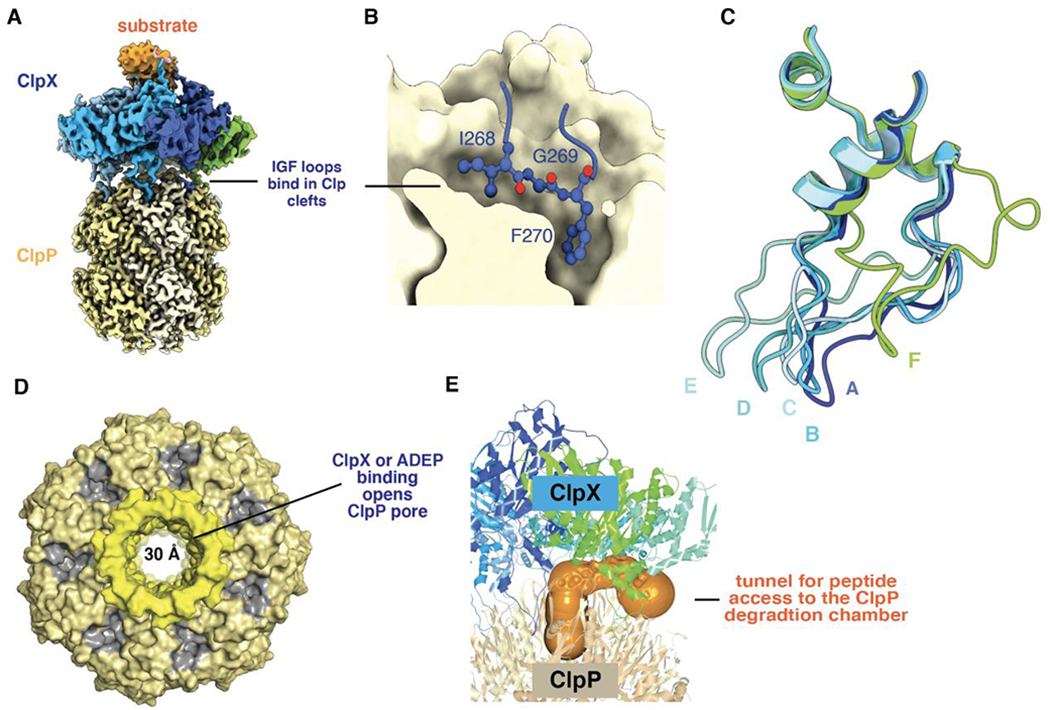
(A) Composite cryo-EM structure of ClpX bound to ClpP and protein substrate (pdb codes 6PPE and 6PP6). (B) An IGF sequence of ClpX binds deeply in a ClpP cleft. (C) IGF loops from aligned ClpX subunits (pdb code 6WRF) adopt a wide variety of conformations with respect to ClpP. This loop flexibility allows ClpX and ClpP to remain stably bound as ClpX adopts different conformations during its ATP-fueled mechanical cycle. (D) Axial view of ClpP (pdb code 6PPE) bound to ClpX (not shown) showing an open axial pore, the clefts that serve as docking sites for the IGF loops of ClpX or ADEPs (colored gray), and the collar of β hairpins that surround the axial pore (colored lighter yellow). (E) The orange surface shows a tunnel, calculated using CAVER (Pavelka et al., 2016), which may allow peptides to enter and leave the ClpP degradation chamber by passing between neighboring IGF loops of ClpX.
