Abstract
Giant unilamellar vesicles (GUVs) are cell-sized aqueous compartments enclosed by a phospholipid bilayer. Due to their cell-mimicking properties, GUVs have become a widespread experimental tool in synthetic biology to study membrane properties and cellular processes. In stark contrast to the experimental progress, quantitative analysis of GUV microscopy images has received much less attention. Currently, most analysis is performed either manually or with custom-made scripts, which makes analysis time-consuming and results difficult to compare across studies. To make quantitative GUV analysis accessible and fast, we present DisGUVery, an open-source, versatile software that encapsulates multiple algorithms for automated detection and analysis of GUVs in microscopy images. With a performance analysis, we demonstrate that DisGUVery’s three vesicle detection modules successfully identify GUVs in images obtained with a wide range of imaging sources, in various typical GUV experiments. Multiple predefined analysis modules allow the user to extract properties such as membrane fluorescence, vesicle shape, and internal fluorescence from large populations. A new membrane segmentation algorithm facilitates spatial fluorescence analysis of nonspherical vesicles. Altogether, DisGUVery provides an accessible tool to enable high-throughput automated analysis of GUVs, and thereby to promote quantitative data analysis in synthetic cell research.
Keywords: giant unilamellar vesicles, image analysis, object detection, open-source software, bottom-up reconstitution, synthetic cell
1. Introduction
Giant unilamellar vesicles (GUVs) are aqueous compartments enclosed by a lipid bilayer membrane.1 Since their diameter is typically between 5 and 100 μm, which is comparable to the size of eukaryotic cells, and their membrane is composed of phospholipids just like plasma membranes, GUVs are considered a good model system for living cells. As such, GUVs have gained great interest from researchers in biochemistry, biophysics, synthetic biology, and applied medicine.
One of the most classical applications of GUVs is in studying the physicochemical properties of biological membranes. Being larger than other biomimetic membrane systems such as large unilamellar vesicles (LUVs) and small unilamellar vesicles (SUVs), GUVs can be easily observed with optical microscopy. Accordingly, GUVs are extensively used to study a wide variety of membrane properties: mechanics,2 lipid diffusivity,3 permeability,4 as well as lipid order5 and domain formation.6,7 Moreover, GUVs are often used to study the biophysical mechanisms that underlie important cellular events such as membrane growth,8 budding,9 fission,10 and fusion.11 Furthermore, GUVs provide a suitable chassis in the endeavor of building synthetic or artificial cells.12−14 In this emerging research field, cellular functionalities are being reconstituted from chemical or biological building blocks with increasing complexity, with the eventual goal to understand the minimal requirements for life at the cellular level.15 Finally, the biocompatibility of GUVs makes them also interesting in the context of targeted drug delivery.16 They overcome the size limitations of the SUVs that are typically used, offering a way to deliver more cargo per particle.
As GUVs are becoming a widely used tool in synthetic biology, also the possibilities for their production are growing. By now, numerous methods have been developed to produce GUVs, ranging from simple and quick bulk methods with low-cost equipment to advanced microfluidic methods. Two major pathways can be distinguished for the production. First, GUVs can be formed by hydration of a dried lipid film, either by spontaneous swelling on solid supports or porous substrates or by application of an electric field.1,17−19 Second, GUVs can be templated from water-in-oil emulsion droplets, for example, by the inverse emulsion method,20,21 with microfluidics22,23 and by continuous droplet interface crossing encapsulation (cDICE).24,25 With the versatile options for formation, the design possibilities for GUVs have become legion: from simple membranes composed of a single lipid type to complex biological lipid extracts,26,27 charged28,29 or bio-functionalized membranes,30 membranes with asymmetric leaflets,31 or including membrane proteins,32 in physiological buffers,29,33 encapsulating functional proteins,8,25 or even active matter.34,35
By far the most widely used characterization technique for GUVs is optical microscopy. GUVs can be imaged in bright-field or by fluorescence microscopy upon inclusion of dyes, either membrane-bound or encapsulated. While most studies with GUVs involve simple wide-field or confocal microscopy, also superresolution microscopy36 and bulk analysis with multiwell plate assays21 and fluorescence-activated single cell sorting (FACS)37 have been employed. For all of the GUV applications described above, it is crucial to evaluate the quality of the produced GUV samples because the success of GUV formation and the resulting vesicle properties can vary substantially dependent on experimental conditions. GUV analysis comes itself with the challenge that in most reconstitution experiments, the formed vesicles are polydisperse in size, shape, the presence of membrane structures, and encapsulated content. The complex appearance of heterogeneous GUV populations therefore demands a quantitative characterization by accurate descriptors and robust statistics.
Despite the experimental ease of producing and imaging GUVs, their quantitative image analysis has received relatively little attention.38 Typically, GUVs are either manually detected in the image and afterward (manually) processed to extract data,30,39−44 or custom-made scripts are used to process specific data sets and generate a predefined set of output parameters.21,25,45−47 While general image analysis software (e.g., ImageJ) can be used with GUV microscopy data, there are no dedicated modules or plugins for GUV detection and analysis, requiring for the user to adapt the built-in methods and, in case of automated analysis, programming knowledge is often needed. Consequently, GUV image analysis is currently time-consuming and nonstandardized, making it difficult to directly compare the outcome of different studies.
In the field of cell biology, analysis workflows do exist for the automated characterization of cell or tissue image data, combining standardized detection modules with reporting a multitude of output variables (reviewed in ref (48)). Unfortunately, these analysis workflows offer limited compatibility with GUV data sets. While cells generally have a complex morphology, GUVs are typically near-spherical, highly symmetric three-dimensional (3D) objects. Due to their often predictable shapes and intensity profiles, rapid and efficient detection and characterization of vesicles benefit from a simplified approach. Furthermore, irrespective of the application in which GUVs are used, the same set of descriptors are typically of interest, in particular vesicle size, shape, membrane intensity (lamellarity), and spatial intensity profiles of GUV membrane and content.
Some examples of openly available softwares dedicated to GUV analysis do exist, laying the ground to make large-scale GUV analysis more accessible. However, they are all either geared toward specific, predefined analysis (membrane permeability,49 heterogeneity in membrane signal,50 pore formation,51 phase separation52) or they have limited compatibility with input data sets and vesicle types (confocal microscopy images,46 spherical vesicles53), requiring a high signal-to-noise ratio of the membrane or predefined vesicle shapes for vesicle detection. In addition, most available software lacks a user-friendly interface that allows for interactivity during the detection and analysis procedures, in turn imposing a steep learning curve on new users.
To meet the requirement for accessible and flexible quantitative vesicle analysis, we have developed DisGUVery, an open-source software for the analysis of GUVs in microscopy images. Our tool encapsulates multiple algorithms for the detection of vesicles and the subsequent analysis of their morphology and content under a graphical user interface (GUI) based on Python. The software is designed to allow for maximal flexibility in data input, processing, and analysis, enabling the user to work with a variety of imaging sources, to export variables of interest at any point during the processing, and to choose between a set of predefined detection and analysis modules. Our toolbox provides a general, fast, and user-friendly approach toward quantitative and high-throughput GUV sample characterization, which should be of broad use in the fields of membrane biophysics, cell biology, and bottom-up synthetic biology.
2. Results and Discussion
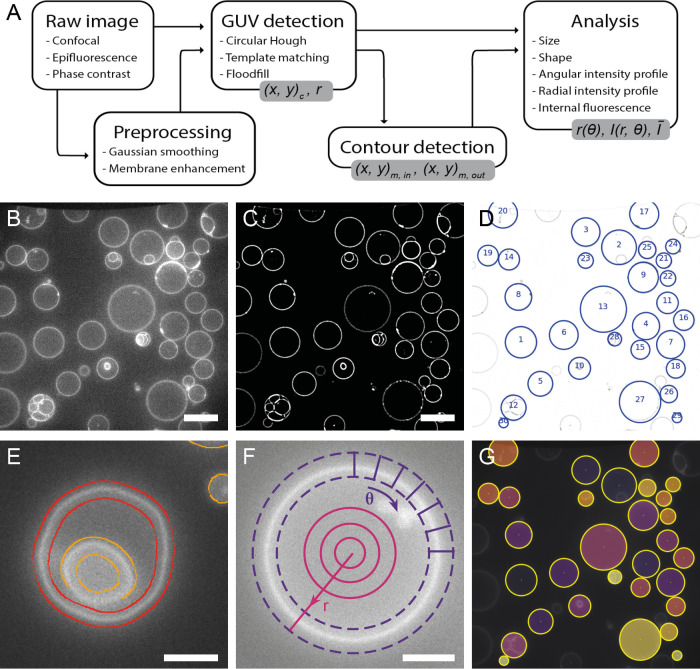
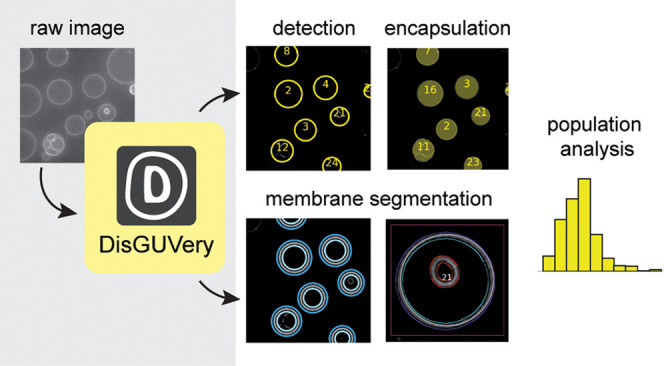
The general workflow of DisGUVery is summarized in Figure 1A. The starting point is a microscopy image with GUVs visible either via fluorescent labeling (wide-field fluorescence or confocal microscopy as shown in Figure 1B) or in phase contrast microscopy. In case of multichannel images, it is possible to select the channel that should be used for GUV detection, typically, but not limited to, a channel where a membrane dye is imaged, or a phase contrast channel in which vesicles are clearly visible. To allow for high-throughput analysis of large GUV data sets, images can be processed in batches both for vesicle detection and subsequent analysis. When processing is done, detection results can be inspected in the software, and erroneously detected vesicles or unsuccessfully processed images can be discarded before further analysis.
Figure 1.
General workflow of GUV detection and analysis by DisGUVery. (A) Visual representation of the workflow. Output variables are shown in gray boxes. (B) Example of an unprocessed single-plane confocal fluorescence microscopy image of GUVs, used as input for the analysis. Scale bar is 20 μm. (C) Processed image after enhancement of the membrane signal. (D) Vesicles detected by circular Hough detection indicated with blue circles and object index number. Contrast is inverted for visualization. (E) Refined contour detection distinguishes the enclosing membrane of a detected vesicle (red) from other fluorescent structures in the image (orange). Scale bar is 5 μm (F) Radial (magenta) or angular (purple) intensity profiles can be extracted from detected GUVs. Scale bar is 5 μm. (G) Masks (colored) can be created from detected vesicles (yellow circles) to extract internal fluorescence of vesicles.
Prior to vesicle detection, background noise can be reduced by processing the input image using a Gaussian smoothing filter followed by an enhancement of the membrane signal (Figure 1C), both of which can be tuned according to the input image. After this optional preprocessing of the image, GUV detection can be done using one of three different methods, all of which yield the indexed locations and sizes of detected GUVs (Figure 1D). To optimize detection, input parameters can be varied in the GUI and the results can be directly inspected, and wrongly assigned vesicles can be manually discarded. Size distributions can at this point be directly computed and visualized, or other, more complex analysis can be pursued.
For many analysis purposes, such as obtaining vesicle shape descriptors of deformed GUVs40 or probing membrane colocalization of fluorescent proteins,54 precise membrane location is required. We have implemented a membrane segmentation algorithm that can be applied to nonspherical vesicles, effectively detecting both the inner and outer edge of the membrane intensity signal (Figure 1E). When high spatial accuracy is not required, users can also make use of the computationally cheaper basic membrane analysis (BMA) feature, where the contour from vesicle detection is simply expanded with a certain width to create a ring that contains the membrane (Figure 1F, purple dashed lines). Regardless of the chosen method for membrane segmentation, the angular intensity profiles and the radially integrated intensity profiles can be extracted (Figure 1F), for example, to retrieve the angular profile of membrane fluorescence,55,56 the angular profile of a fluorescent membrane-binding molecule,57 or to quantify membrane localization of an encapsulated molecule30 or an externally added membrane-binding protein.8
Besides the radial and angular intensity profiles, which provide information about the spatial distribution of fluorescent probes, the average intensity of the vesicle lumen can also be extracted for detected vesicles using the Encapsulation Analysis module (Figure 1G). Analysis of internal fluorescence is essential for studying the efficiency of encapsulation of molecules and other components,25,58 for permeabilization assays where transport of a fluorescent probe across the membrane is tested,59−61 and for fluorescence-based measurements of the activity of internal metabolic pathways.8,62
The analyses mentioned above are some of the methods that we have predefined in the software. However, we want to stress that the use of DisGUVery is not limited to these analyses. Since it is possible to export results from object detection, contour detection, and from the analyses at any point in the process, users can extract the relevant information and perform their own analyses outside the software.
2.1. Vesicle Detection
We have implemented three different methods for the detection of vesicles in microscopy images: circular Hough transform (CHT), multiscale template matching (MTM), and floodfill detection (FF). As the underlying principle for object detection is different for each of the methods, they allow detection of a variety of vesicle shapes and imaging sources. The first method, based on the circular Hough transform of the object edges,63 is commonly used in the detection of GUVs as it recognizes circular objects with little influence on the intensity profile.49,53 As a result, detection by CHT depends mostly on vesicle shape and not on image intensity, providing a robust method with a high selectivity toward circular vesicles. When the vesicle shape is not circular, but is predictable, for example, by having a population of similar-looking vesicles in an image, detection can be done via the second method: template matching.64 We have implemented a slight variation of this method, Multiscale Template Matching (MTM), by allowing the rescaling of the template to multiple sizes. MTM works by the convolution of the image with a target object, or template, which can be an image of a typical vesicle. Regions in the image are then assigned as detected objects when this template matches the region, with the scaling of the template enabling the size-invariant detection of vesicles. The third method, Floodfill detection (FF), is based on an absolute intensity difference between membrane and background signals.65 By thresholding the image, membranes can be distinguished from the background and closed membrane contours in the thresholded image are assigned as vesicles. Floodfill detects vesicles based on membrane fluorescence, regardless of their shape. Note that FF has been implemented previously for vesicle detection by Blanken et al.,46 but with a different starting point for the floodfill algorithm (the seed point). While their algorithm floods all of the regions within GUVs by scanning a range of intensity thresholds and seed points, ours floods the surrounding background, which has the computational advantage of using only a single thresholding intensity and a single seed point.
We evaluated the performance of the three vesicle detectors on different types of microscopy images: fluorescence confocal, wide-field fluorescence, and phase contrast. We focused on two main aspects to determine the quality of the detectors: how good are they at detecting vesicles within an image, and, how sensitive is this detection to different factors, e.g., detector parameters or image source. Detection outcomes of the software were benchmarked against human visual detection. We started by optimizing the detector parameters on a single image. Figure 2A–C shows an example of the detection results for all detectors on a single confocal image. In this case, the optimal parameters are those which allow the detection of the highest number of vesicles in the image, regardless of their characteristics, while avoiding artifacts in the detection. Once this optimization has been done, we explore the parameter space of each of the detectors and evaluate their detection accuracy using the F1 score,66 defined as:
| 1 |
Here, 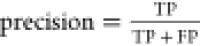 and
and 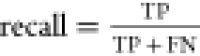 with TP, FP, and FN being true positives,
false positives, and false negatives, respectively. In this study,
true positives are detection results that correspond to vesicles,
false positives are identified objects that are not vesicles, and
false negatives represent GUVs that have not been detected. The reference
human visual analysis was performed by a single observer by counting
all GUVs in the images, regardless of vesicle size, appearance, or
location in the image. GUVs at the edge of the image were included
as long as a part of the membrane was visible.
with TP, FP, and FN being true positives,
false positives, and false negatives, respectively. In this study,
true positives are detection results that correspond to vesicles,
false positives are identified objects that are not vesicles, and
false negatives represent GUVs that have not been detected. The reference
human visual analysis was performed by a single observer by counting
all GUVs in the images, regardless of vesicle size, appearance, or
location in the image. GUVs at the edge of the image were included
as long as a part of the membrane was visible.
Figure 2.
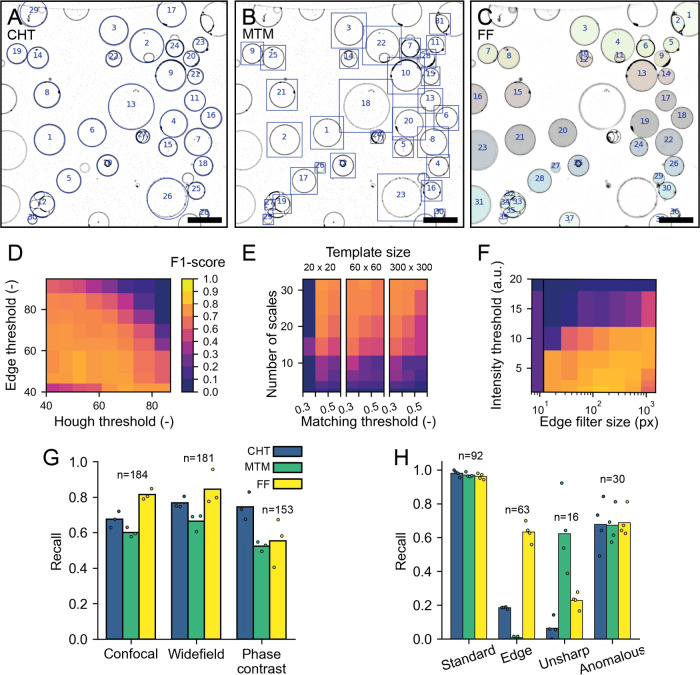
Vesicle detection results. (A–C) Detected vesicles with the circular Hough transform (A, blue circles), multiscale template matching (B, blue bounding boxes), and floodfill (C, colored objects). The contrast of the images has been inverted for visualization purposes. Scale is 20 μm in all images. (D–F) F1 score of CHT (D), MTM (E), and FF (F) for different parameter values (see methods for details on the parameters). The color scale in (E, F) is the same as in (D). (G, H) Performance of vesicle detectors. Individual data points represent results of the human individual observers, and bars represent average recall values; n is the number of vesicles per category. (G) Recall of vesicle detection for confocal fluorescence, wide-field fluorescence, and phase contrast images using the three different detectors. (H) Detection recall for different subcategories of vesicles in confocal images. See the main text for a more detailed explanation of the use of categories.
We have chosen to use the F1 score as an output metric to evaluate our detectors because it is mainly penalized by false negatives and false positives, both of which are useful output parameters in object detection. As such, F1 is amply used in object detection problems.67 Since the F1 score is the harmonic mean of precision and recall, both are weighed equally into a single output. While recall represents the fraction of objects in the image that are detected, precision denotes which fraction of detected objects are vesicles. Figure 2D–F shows the F1 scores as a function of pairs of critical parameters inherent to each of the detectors, with the exception of Floodfill (FF). For FF detection, we chose instead the size of the filter for membrane signal enhancing in the preprocessing step, as we have found it to be critical for the method performance (Figure 2F). We find that all detectors show a region within their parameter space in which the F1 score is maximum and their performance is best. Note that the F1 score only changes within 10% of its maximum value for a large set of parameters, suggesting that a precise optimization of the parameters is not necessary, which facilitates batch-processing of data sets with similar images. Notably, the performance of CHT detection drops only at large input thresholds (∼50% increase from the optimal value) for both input parameters (Figure 2D), reflecting its high tolerance to nonoptimal parameters. For MTM, the number of scales used to resize the template is the critical factor in achieving good performance. This is strongly dependent on the data set used, more specifically on the polydispersity in vesicle sizes. For the wide distribution of vesicle sizes in our sample, a large number of template scales allows precise matching of the template across the entire size range. To ensure that the size of the template prior to scaling does not play a crucial role in detection, for example, due to pixelation effects, we tested templates of different sizes, which resulted in similar performance (Figure 2E). As expected, FF detection depends greatly on the intensity threshold (Figure 2F), with higher values not allowing the vesicles to be properly segmented in the binary mask of the thresholded image. Interestingly, in the images tested, the preprocessing step of membrane enhancement is crucial for FF detection to succeed, as without it, FF detection simply fails to detect vesicles as is demonstrated in Figure 2F at the smallest edge filter size.
We then investigated the extent to which the imaging conditions impact the detection methods, for example, by changing the type of microscopy used to visualize the vesicles. We compiled a data set of five images for each one of the three standard microscopy techniques mentioned above, resulting in a total of over 200 vesicles for each imaging method. For each data set, we perform the detection using the parameters that were fine-tuned for a random image within the set. To measure the performance of the detectors, we evaluate separately precision and recall. The reason for this split being that all detectors show a high precision (between 0.9 and 0.99) for the different data sets (Figure S1), with any differences in the detection performance being represented predominantly in the recall metric. In Figure 2G, it can be seen that in both confocal fluorescence and wide-field fluorescence images, all detectors are able to detect vesicles with a recall between 0.6 and 0.8, meaning that 60–80% of vesicles are properly detected. However, for phase contrast images, recall decreases for MTM and FF detection (to 50%). The low performance of the FF detector in phase contrast is expected based on the intensity profile that vesicles show in this type of microscopy, where the GUV membrane does not represent an intensity maximum but instead the steepest intensity gradient. Furthermore, intensity variations, either due to inhomogeneity of illumination, out-of-focus light or due to the presence of surrounding objects, interfere with detection with both MTM and FF. Unsurprisingly, CHT is the most robust method across imaging sources for the sample we investigated, emphasizing its stronger dependence on vesicle shape rather than on intensity profile. While we have only tested detection with these three imaging types, we anticipate that DisGUVery is also compatible with differential interference contrast (DIC) and bright-field microscopy, provided that the vesicles are clearly visible.
Given that the detectors target different types of objects, we next looked into what kind of vesicles were being detected by each method. We therefore manually divided all vesicles analyzed in Figure 2G into four categories: vesicles that were located at the edge of the image (“edge”), vesicles that were out of focus (“unsharp”), anomalous vesicles (“anomalous”), and vesicles that do not belong to any of the first three categories which we called “standard” vesicles. A gallery of example vesicles for all subcategories can be found in Figure S2. Anomalous vesicles could be vesicles with a bright membrane signal in their lumen, with very heterogeneous membrane signal, or vesicles that were stuck together in aggregates. Typically, the standard vesicles are those needed for further analysis. In Figure 2H, we can clearly see that while standard vesicles are detected similarly by all methods, with recall being nearly 1.0, vesicles on the edge of the image and those that are out of focus can be easily detected or filtered out depending on the method of choice. CHT and MTM filter out most vesicles at the edge. Furthermore, CHT also misses the unsharp vesicles, which are not detected by FF either. Interestingly, all detectors perform similarly for the anomalous vesicles, with more than 60% of them being detected. Together, these results show the robustness of the different detectors, allowing detection to be performed on a variety of vesicle types and imaging sources.
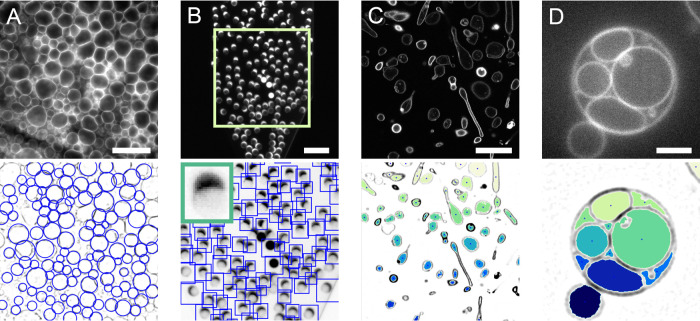
To illustrate the range of applications of the three detectors for synthetic cell research, we performed vesicle detection in a selection of proof-of-principle experiments (Figure 3). First, we investigated if our methods could be used to track GUV size and number during swelling-based GUV formation. GUV swelling methods include electroformation and gel-assisted swelling and are by far the most popular formation methods, as they are fast and easy and yield large numbers of GUVs.19,38,68,69 In these experiments, lipids are first dried on a surface, which can be a hydrogel, an electrode, glass, Teflon, or a porous material. Subsequent addition of a swelling solution leads to swelling of the lipid film and formation of large numbers of GUVs that are closely packed above the swelling surface (Figure 3A, top). Using CHT detection, we efficiently detect spherical vesicles even at high surface coverage (Figure 3A, bottom). By automated detection of GUVs during their formation, growth kinetics could easily be obtained. Since detection by CHT relies on vesicle shape rather than intensity, the method is largely insensitive to touching vesicles or high background fluorescence, both of which are more likely at high packing density. Furthermore, the ability to specify a minimum and maximum GUV radius prevents detection of false positives in dense samples. In addition to GUV production by swelling, detection at high packing density is also relevant for studies on GUV–GUV adhesion70 or while building multibody GUV tissues.71
Figure 3.
Applications of vesicle detection methods. Images at the top are the input fluorescence images, and images at the bottom show the detection results. Contrast has been inverted for detection results to improve visualization. (A) GUVs growing on top of a hydrogel following the gel-assisted swelling method are detected with CHT. Scale is 40 μm. (B) Microfluidic production of GUVs imaged with a low-magnification objective. MTM is employed to detect GUVs in the microfluidic channel. Produced vesicles contain a lipid-rich octanol pocket, visible as a bright cap. Scale is 100 μm. Inset in the bottom image is the template used for detection. (C) Encapsulation of stiff actin bundles in GUVs leads to deformation of the vesicles (data by F.C. Tsai, from ref (40)). The nonspherical vesicles can be detected with FF detection. Scale is 20 μm. (D) GUV formed by gel-assisted swelling containing large internal vesicles. FF detection on this multivesicular GUV leads to the detection of the compartments rather than detection of the enclosing GUV. Scale is 5 μm.
As an alternative to the classical swelling methods, microfluidic vesicle production is becoming increasingly popular, with multiple new techniques being published yearly (reviewed in refs (72, 73)). Microfluidics offers superb control over vesicle formation, making it a powerful tool in the synthetic cell engineering field. Vesicles can be imaged in situ as they are being produced on-chip, using objectives with a large working distance with low magnification. This typically yields low-resolution images of vesicles. In line with MTM’s ability to detect out-of-focus vesicles (Figure 2H), MTM also proves to be suitable for GUV detection in low-resolution images of microfluidic GUV fabrication, as we demonstrate with an octanol-assisted liposome assembly (OLA) experiment22 (Figure 3B). Detection of vesicles on-chip enables users to extract GUV production rates and corresponding size distributions in microfluidic experiments. MTM detection does not require a sharp outline of the vesicle, but only a template that resembles the vesicles that need to be detected. Since the template can easily be picked from the image itself, MTM provides a versatile tool for vesicle detection even in low-resolution images. As an alternative to MTM, CHT is also a useful detector in microfluidic experiments (Figure S3) since production by microfluidics often leads to spherical vesicles at high packing density with a narrow size distribution.58
While CHT and MTM are both shape-sensitive detectors, FF can detect vesicles of any shape. Having a detector that does not rely on vesicle shape is valuable, as shape control and GUV deformation are essential aspects of synthetic cell engineering.9,40,74,75 We demonstrate the use of FF on GUVs deformed by encapsulated stiff actin bundles (Figure 3C). In this experiment, filamentous actin is co-encapsulated with the bundling protein fascin, resulting in the formation of actin bundles up to tens of micrometers long.40 Due to the high stiffness of these bundles, GUVs are deformed, resulting in elongated vesicles and actin-filled membrane protrusions. In Figure 3C, it can be seen that FF detects all vesicles irrespective of shape. In turn, detection by FF can be used as a starting point for vesicle deformation studies. Another application of the FF detector is found in the image segmentation of GUV internal compartments. Similar to cells, GUVs can be compartmentalized to spatially separate cellular processes.76,77 Compartmentalization is becoming more popular in synthetic cell research, as distinct reaction environments are desired for reconstitution of increasingly complex processes. Due to its shape-insensitive detection, the FF method is suitable for detecting GUV compartments with random shapes and sizes as illustrated in Figure 3D. In this way, detection of compartments could be used to monitor the internal activity of cellular processes.
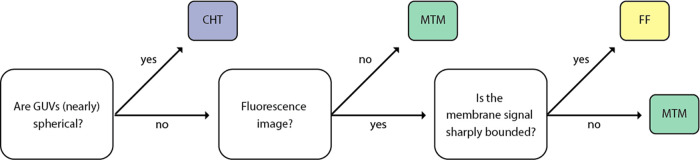
Altogether, the three vesicle detection methods make DisGUVery useful for a wide range of synthetic cell research applications. We note that for most images, more than one detector can be used (illustrated in Figure S3). Which detector is preferred will depend on the input image type, sample appearance, and desired analysis (Figure 4).
Figure 4.
Decision tree for choosing one of DisGUVery’s detection modules based on sample and image properties.
2.2. Membrane Analysis
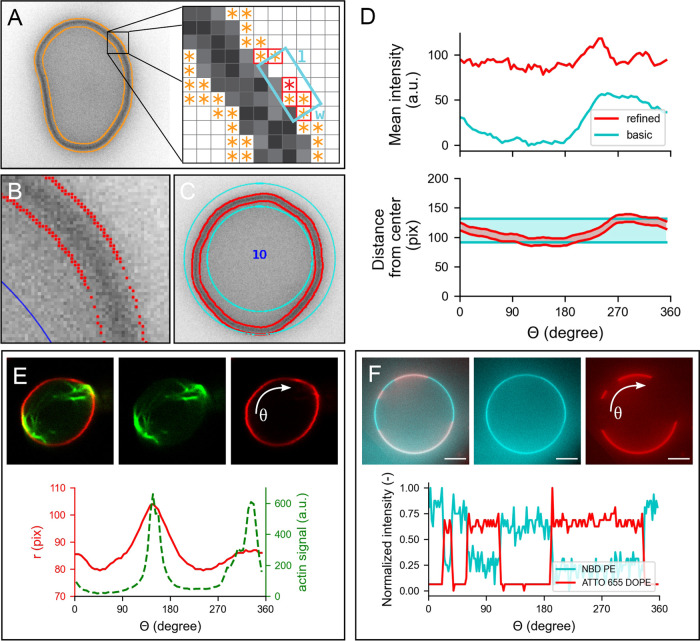
Membrane detection and analysis are important for a wide range of GUV studies, as they allow vesicle shape characterization and quantitative analysis of the membrane fluorescence and/or membrane-binding proteins or other molecules. We have implemented two modules to perform membrane analysis on the detected vesicles: refined membrane detection (RMD) and basic membrane analysis (BMA). We have developed RMD to enable the tracking of the membrane contours, facilitating the capture and quantification of global and local deformations. This method is based on a modified Canny edge detector78 that we have combined with a directional search algorithm to assign the detected edges to the inner and outer contours of the membrane (Figure 5A–C, contours shown in red). While the position of the detected contour can be affected by the choice of kernel size used with the edge detector (a predictable offset is introduced), the membrane position, taken as the midpoint between the inner and outer contour, will remain independent of the kernel size for confocal fluorescence images. Compared to RMD, BMA is a faster and simpler method to analyze the vesicle fluorescence signal, but at the expense of lower spatial accuracy. In BMA, a region of interest (ROI) is created by a simple expansion of user-defined width around the boundaries of the detected vesicle. In case of CHT detection, this results in a circular ring as shown in Figure 5C.
Figure 5.
Membrane analysis by DisGUVery. (A) Schematic overview of the refined detection method. Left: wide-field fluorescence image (inverted) of a GUV with all detected edges (orange). Right: edge points (orange) within the search box (cyan) of size l × w are connected (red border). (B) Zoom-in on membrane edges detected by refined detection (red), displayed on top of the inverted wide-field fluorescence image of a GUV. (C) Segmentation of the membrane area as defined by basic membrane analysis (cyan) and refined membrane detection (red). (D) Angular profile of membrane properties from the vesicle in (C) extracted by basic membrane analysis (blue) and by refined membrane detection (red). Top: mean intensity per angular slice with an angular separation of 5°, and a ring width of 30 pixels for BMA. Bottom: radial distance to inner and outer boundaries from the center of the vesicle. (E) Refined membrane detection on a nonspherical GUV deformed by actin bundles. Insets: composite confocal image of a GUV membrane (red) deformed by actin-fascin bundles (green) (data by F.C. Tsai, from Tsai et al.40). Plot: angular profile of the membrane’s radial distance (red) and integrated actin intensity (green). (F) Basic membrane analysis of a phase-separated membrane containing DOPC:DPPC:cholesterol:NBD-DPPE:ATTO655-DOPE in a 31.8:48:20:0.1:0.1 molar ratio. Insets: image of vesicle labeled with NBD-DPPE (blue) and ATTO655-DOPE (red). Plot: angular profile of both dyes extracted by basic membrane analysis, normalized to unity by subtraction of the minimum signal followed by division by the remaining maximum signal.
After membrane segmentation, either by contour detection in RMD or by defining a region of interest in BMA, it is possible to extract the angular and radial intensity profiles of the entire vesicle. The intensity profiles are calculated by creating angular or radial slices, of size Δθ or Δr, and computing the corresponding intensity metrics for each slice, thus taking into account all intensity values of the detected vesicle and reducing the effects of discretization associated with single linear profile extraction. For the vesicle in Figure 5C, the angular intensity profile using the mean membrane intensity from each angular slice is shown in Figure 5D (top). Note that, although the trend of the mean intensity profiles is similar for RMD and BMA, the values differ greatly. This is a consequence of the wider segmentation ring of BMA (Figure 5D, bottom), which, when used to compute the mean intensity values, introduces the influence of the background signal, unlike the contained segmentation done by RMD. When required, an appropriate background correction that is in-built in the software can be used to minimize artifacts induced by the width of the BMA segmentation ring (see Figure S4). Furthermore, it can be seen that the BMA profile shows a fluorescence increase from θ = 250 to 300°, while this effect is much weaker for RMD. We attribute this apparent increase in fluorescence to the fact that the membrane shows an outward deformation around θ = 250° (Figure 5D, bottom), causing a larger part of the BMA slices starting from that angle to be filled with membrane compared to other slices. Since in RMD the ROI always tightly confines the membrane, extraction of fluorescence intensity is much less sensitive to membrane shape. Dependent on the nature of the data and the required analysis, the choice of descriptive metrics can have a significant contribution of imaging artifacts or other sample-related noise (see the Supporting Information, SI). For example, while both the integrated intensity and the mean intensity are influenced by the background signal, the latter will also depend on the number of pixels within the slice. Polydisperse samples, where there is a large variation in vesicle size, will thus require a careful interpretation of the results and likely, a different metric to analyze the data compared to more monodisperse samples.
To further illustrate the applicability of both methods in the quantitative characterization of GUV membranes, we show how RMD can be used to analyze the membrane and content of a deformed vesicle (Figure 5E), while we use BMA for an example on phase-separated membranes (Figure 5F). In the first example, a GUV is deformed to a prolate shape by encapsulated filamentous actin that is bundled by the bundling agent fascin.40 We used RMD to track the membrane contour position, and additionally, we extracted the angular profile for the average actin intensity from the RMD contour (Figure 5E, bottom). The plot clearly shows two peaks in membrane position around angles 150 and 340°, which correspond with the peaks in actin intensity. If desired, the obtained contour coordinates can be exported to compute other shape descriptors of interest. In this way, membrane deformation by fluorescent structures can be quantified in an automated way for vesicle populations, enabling an accessible and quantitative approach in GUV deformation studies. Besides its use in actin-mediated GUV deformation studies,40,44,79−81 this analysis is also valuable in other studies on global vesicle shape deformation, for example by other proteins involved in cytokinesis such as the bacterial division proteins FtsZ82 and Min system,74 by other membrane-binding proteins,9 DNA origami,83 by microfluidic traps,75,84 or by spontaneous membrane fluctuations.85 Furthermore, RMD could be applied to characterize local membrane deformations, such as protrusions86 or nanotubes.28,55
In cases where vesicles are rather spherical and their shape well characterized, BMA is a useful tool to study the fluorescence signal of the GUV membrane. In Figure 5F, we show a GUV composed of a lipid mix of DOPC:DPPC:cholesterol:NBD-DPPE:ATTO655-DOPE in a molar ratio of 31.8:48:20:0.1:0.1. In this ratio, the lipids form two spatially separated phases:87 a liquid-ordered phase containing mainly DPPC lipids and cholesterol, and a liquid-disordered phase containing mainly DOPC lipids. While NBD-DPPE partitions preferentially into the liquid-ordered phase (blue), ATTO655-DOPE accumulates in the liquid-disordered phase (red). Using CHT detection followed by BMA, we obtained the angular profiles of both membrane dyes. The intensity profiles indeed clearly show that both dyes have a preferential presence in either one of the two phases. While here we show the example analysis for one single GUV, we would like to stress that BMA performs membrane analysis at high computation speed, enabling the analysis of membrane fluorescence for hundreds of vesicles within minutes. Next to lipid-lipid phase separation studies, BMA could be used for membrane quenching experiments,88 to probe the homogeneity of a reconstituted actin cortex,30 or for analysis of spectral images in lipid packing studies using polarity-sensitive probes.89
2.3. Population Analysis
So far, we have demonstrated DisGUVery’s working principles and the performance of detection and membrane analysis on single vesicles or single images. However, for the analysis of GUV experiments, it is often desired to analyze large numbers or time-lapse series of vesicles. We implemented a batch-processing option that allows for the semiautomated analysis of multiple images, making population characterization on large data sets accessible and enabling easy identification of statistical differences. We illustrate the potential of the batch-processing feature with two quantitative analyses: binding of small vesicles to GUVs using membrane-anchored oligonucleotides and the encapsulation of a fluorescent protein inside GUVs.
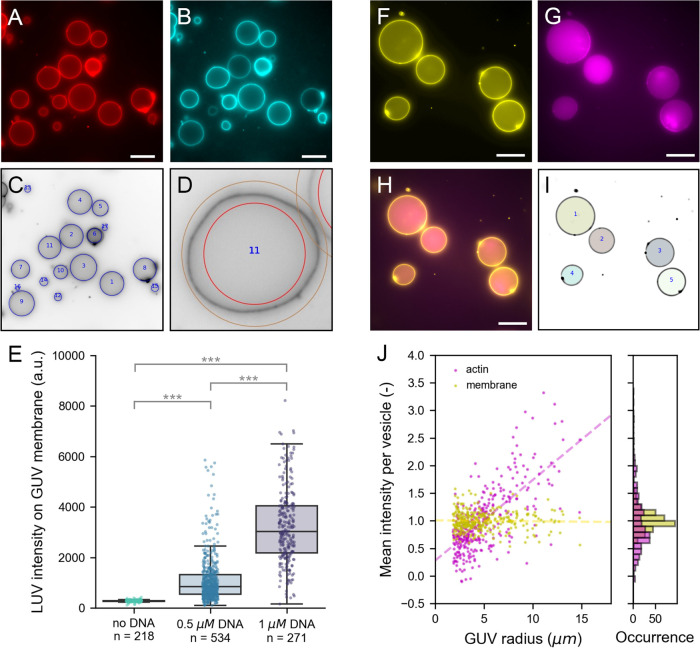
In the first case, we utilize the membrane analysis module on a population of vesicles where we bound large unilamellar vesicles (LUVs) to GUVs using membrane-anchored oligonucleotides (Figure 6A–E).90,91 While GUVs have diameters of tens of microns (Figure 6A), the LUVs used in this study have a diameter of approximately 200 nm, close to the size of the diffraction limit. To generate specific binding between GUVs and LUVs, we incorporated one type of single-stranded DNA (ssDNA) in the GUVs and the complementary ssDNA in LUVs. Here, we set out to test if the extent of LUV-GUV binding could be regulated by varying the DNA concentration. Therefore, we incubated vesicles with 0.5 μM DNA, 1 μM DNA, or no DNA at all prior to mixing LUVs with GUVs. LUVs were doped with a fluorescently tagged phospholipid for visualization and quantification. To allow vesicle detection that is not biased by LUV binding, we independently labeled GUV membranes with another fluorescent phospholipid. When both types of vesicles were incubated with 1 μM of the complementary DNA strands prior to mixing, LUVs clearly localized on the GUV membranes (Figures 6B and S5A), while we observed no colocalization in the absence of DNA (Figure S5B). To quantify LUV binding, we first detected GUVs in the Atto488 channel using CHT detection (Figure 6C). Membrane fluorescence was analyzed using the basic membrane analysis because it is computationally light and our analysis did not demand a high spatial accuracy (Figure 6D). We chose a large (50 pixel) ring width to be able to extract membrane fluorescence also from nonspherical vesicles that were naturally present in the sample. While the software exports multiple intensity metrics from the angular slices, we performed our analysis using the intensity maximum per slice to minimize the effect of the background signal (see the SI). Given that the maximum is sensitive to fluorescence outliers, for example, caused by bright membrane structures or touching vesicles, we finally take the median of all angular maxima to represent the vesicle average. To correct for background intensity, we subtract the radial intensity average just outside the vesicle from the vesicle-average LUV intensity. In this way, we analyzed over 1000 GUVs in 50 different images. The results are plotted in Figure 6E. In the absence of DNA, LUVs do not bind to GUVs, in line with what is seen in the image (Figure S5). Upon DNA addition, membrane analysis shows a clear increase in membrane localization of LUVs. Furthermore, quantitative membrane analysis reveals that the LUV intensity is significantly higher when using 1 μM DNA than 0.5 μM DNA (Student’s t-test, p ≪ 0.001). The data in Figure 6E underlines why population statistics can be essential for analyzing GUV data sets. While vesicles with similar LUV intensity exist in both populations, a statistical difference between the two populations can only be proven when a large number of vesicles is analyzed. In this way, high-throughput membrane analysis helps to quantitatively investigate the effect of experimental parameters on GUV membrane studies.
Figure 6.
Population analysis. (A–E) Analysis of LUVs binding via membrane-anchored oligonucleotides to GUV membranes. (A) Atto 488 DOPE-labeled GUVs produced by gel-assisted swelling. (B) Atto 655 DOPE-labeled LUVs localize on GUV membranes when both are incubated with 1 μM cholesterol-DNA. (C) CHT detection in the Atto488-channel (inverted contrast). Detected vesicles are indicated with blue circles. (D) Example of the detection ring of 50 pixels width used for basic membrane analysis. (E) Bar plot of LUV intensity on the GUV membrane at different DNA concentrations. Each point represents the LUV intensity on an individual vesicle; *** indicates statistically significant difference with p < 0.001. (F–J) Analysis of fluorescent monomeric actin encapsulated in GUVs using cDICE. (F) DOPC GUVs labeled with 0.1% (mol/mol) 18:1 Cy5 PE. (G) Encapsulated actin of which 10% is labeled with Alexa 488. (H) Composite image of membrane and actin. (I) Results of FF detection. Masks represent detected vesicles. (J) Mean intensity normalized by population average of actin (magenta) and membrane (yellow) plotted against the GUV radius (left) and shown in a histogram (right). Dashed lines in the scatter plot are linear regression results for actin (magenta, slope is 0.15) and membrane (yellow, slope is 0.00). All images are wide-field fluorescence images. Scale bar is 20 μm in all images.
In the second example of population analysis, we perform an encapsulation analysis using the Encapsulation Analysis module in DisGUVery. Encapsulation of molecules, proteins, vesicles and even living cells inside GUVs is becoming more and more important as GUV-based reconstitution experiments are increasing in complexity.92,93 Besides controlling which types of molecules end up in the GUVs, also their concentration and stoichiometry often need to be regulated for them to function properly. It is essential to evaluate the quality of encapsulation, as this varies substantially between experiments, depending strongly on the way the GUVs are produced as well as on the molecule that needs to be encapsulated.12,25,94 When the encapsulated molecule can be visualized with fluorescence microscopy, the encapsulation efficiency can be determined as the distribution of internal fluorescence of the encapsulated molecule across the GUV population. To demonstrate this, we encapsulated monomeric actin in GUVs using the continuous droplet interface crossing encapsulation (cDICE) technique24 following the protocol outlined by Van de Cauter et al.25 In this experiment, 10% of the actin monomers were labeled with Alexa 488 to allow for fluorescence visualization. Vesicles were imaged in wide-field fluorescence microscopy to capture the signal of the entire vesicle volume in a single frame. From Figure 6F–H, it can be seen that the actin signal is strongly enhanced inside the GUVs compared to the outer solution and that the observed fluorescence varies among vesicles. To quantify the encapsulation efficiency, we first detected vesicles based on the membrane signal by means of the FF detection method (Figure 6I). The advantage of using FF detection is that detected masks directly match the projected shape of the vesicle lumen, independent of vesicle shape and size. Using DisGUVery’s Encapsulation Analysis module, we then extracted the mean intensity of the mask region that overlays the vesicle lumen for both the actin and membrane signal of each vesicle. Furthermore, in each image, we determined the background signal for each imaging channel by taking the mode of the intensity histogram. Background signals were subtracted from the mean intensity per vesicle to finally yield the corrected mean intensity per vesicle. In total, we analyzed 329 vesicles in 22 images of one preparation. In Figure 6J, we show the distribution of the corrected mean intensities for actin and the membrane. Note that in the wide-field fluorescence imaging mode, the fluorescence emission from the entire focal volume is projected onto the imaging plane. Since the focal depth of the system is larger than the vesicle size, we expect a clear dependency on the vesicle size for any fluorescent molecule distributed in the volume of the GUV. In contrast, the membrane fluorescence signal is localized in the surface area of the vesicle, meaning that the same volume of fluorescent membrane probes is always projected onto the focal plane, independent of vesicle size. Indeed, our analysis at the population level reveals the expected linear trend for the intensity of encapsulated actin as a function of GUV radius (Figure 6J, dashed line) indicating that the projected actin signal of a vesicle depends on the vesicle volume. The membrane mean fluorescence, on the other hand, shows no dependency on the vesicle radius, confirming that the fluorescent probe is membrane-bound. While membrane localization of a fluorescent probe can be obvious from images, the power of population analysis is to be able to reveal this also in data sets where this is less trivial, for instance when the fluorescence intensity between vesicles is heterogeneous, or when the signal-to-noise ratio is lower than in the example. In addition, the ability to quantify this localization is useful in many cases, for example when probing membrane interactions. Interestingly, Figure 6J reveals that the mean intensity spread within a vesicle size range is larger for actin than it is for the membrane, reflecting the variability from protein encapsulation across vesicles. This analysis yields a relative measure of variations in encapsulation efficiency among GUVs, which, once combined with a calibration, could be used to evaluate absolute concentrations inside GUVs.
3. Conclusions
Giant Unilamellar Vesicles have become a widely used system for research in biophysics and synthetic biology. As the versatility and complexity of applications grow, and in concert the number of GUV formation methods, it becomes increasingly important to perform rigorous and standardized quantitative analyses. Here, we presented DisGUVery, an open-source software that we have developed for the high-throughput detection and analysis of GUVs in a wide range of microscopy images.
Since the detection of GUVs is the first step in any type of analysis, we have done an in-depth characterization of the object detection algorithms that we have adapted and implemented. Our results show that each detector can be used as a filter for specific vesicle types and that we are able to overcome the influence of imaging source by careful selection of the detector. By testing and demonstrating detection in a broad range of typical GUV samples, we show that DisGUVery fits in with many areas of GUV research. So far, the simplicity of GUVs combined with our hands-on experience in GUV research has allowed us to develop lightweight algorithms with good detection performance. However, we note that with increased morphological complexity, it might be necessary to use more complex detectors, such as supervised machine learning.95 Even then, our software can serve as an accessible basis for generating training data sets for machine learning, thanks to the automated high-throughput segmentation algorithms.
As many GUV studies rely on the shape and fluorescence of the membrane, we implemented a set of tools for membrane segmentation, which can be chosen depending on the spatial accuracy needed. Notably, we developed a membrane contour tracking method by coupling an edge detector with a directional search algorithm that takes advantage of the unique intensity profile of the membrane fluorescence. Furthermore, we showed how a contained membrane segmentation can easily identify local deformations and be less influenced by the background signal compared to a user-defined ROI that segments the membrane. Nevertheless, we illustrated how even a basic segmentation, in combination with high-throughput analysis, can identify statistical differences between GUV populations. Although we focused here on the intensity of the membrane, note that DisGUVery also allows us to obtain the angular and radial intensity profiles of any imaging channel, allowing the user to study spatial distribution of encapsulated content. Altogether, the membrane analysis modules can be used to extract a wide range of vesicle properties, including GUV shape, internal fluorescence, membrane localization of fluorescent proteins, or formation of internal structures. As such, the software can be used for all sorts of assays, such as membrane permeabilization studies, reconstitution of cytoskeletal networks, microfluidic vesicle production, GUV deformation studies, or membrane fusion assays. Although DisGUVery has been developed originally for the detection and analysis of vesicles, the software might be equally useful for data analysis in other research domains involving similar types of microscopy data, such as colloidal and interfacial science.
Concluding, DisGUVery offers an accessible way to perform fast but thorough quantitative analysis of GUV microscopy images. By combining versatile vesicle detection and analysis algorithms, the software can robustly be employed for any type of GUV research. This makes DisGUVery a powerful tool that can help the field to progress toward more quantitative, population-based research.
4. Materials and Methods
The unprocessed microscopy images were analyzed with DisGUVery, our custom analysis software written in Python. DisGUVery is available from an open-source repository (github.com/DisGUVery) and comes with a user guide (see the SI) that describes the program step-by-step using snapshots of intermediate steps in the image processing pipeline, as well as a clear description of the accepted image files and data generated by the software. Further documentation and a data set for testing can also be found in the repository. All data and postprocessing scripts as discussed in this paper are available upon request.
4.1. Experimental Data
4.1.1. Chemicals and Proteins
From Avanti Polar Lipids, we obtained the lipids l-α-phosphatidylcholine (eggPC), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-distearoyl-sn-glycero-3-phospho-ethanolamine-N-[methoxy(poly(ethylene glycol))-2000] (PEG2000-DOPE), 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(Cyanine 5) (Cy5-DOPE), 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxa diazol-4-yl) (NBD-DPPE), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Rhodamine-DOPE). The lipids ATTO 488 DOPE and ATTO 655 DOPE were obtained from ATTO-TEC GmbH (Siegen, Germany). All lipids were stored in chloroform at -20 °C under argon. The chemicals d-(+)-glucose, sucrose, Tris-HCl, KCl, 1-octanol, glycerol, Poloxamer 188, cholesterol, dithiothreitol (DTT), protocatechuic acid (PCA), and the proteins protocatechuate dioxygenase (PCD) and β-casein were obtained from Sigma-Aldrich. For gel swelling, we used poly(vinyl alcohol) (PVA) of 145 kDa, 98% hydrolyzed, obtained from VWR, Amsterdam, the Netherlands. Actin for the encapsulation experiments was purified in-house as described in ref (96). Alexa-488 labeling of actin was done in-house following ref (97).
4.1.2. GUVs for Detection
All GUV images (except Figures 3C and 5E, as described below) analyzed in this work were obtained using an inverted microscope (Nikon Ti Eclipse) with a digital CMOS camera (Orca Flash 4.0). Imaging chambers were passivated with β-casein to prevent membrane adhesion.
The vesicles that were used to illustrate DisGUVery’s workflow (Figure 1A), to test detection (Figure 2), the high-density GUVs growing on top of a hydrogel (Figure 3A) and the compartmentalized vesicle (Figure 3D) were produced by poly(vinyl alcohol) (PVA)-assisted swelling following ref (19). Membranes typically consisted of 99.9% EggPC lipids and 0.01% fluorescent ATTO 655 DOPE. GUVs were typically produced in a swelling solution containing 200 mOsm sucrose and 10 mM Tris-HCl at pH 7.4, and imaged in an isotonic glucose solution (200 mOsm glucose, 10 mM Tris-HCl at pH 7.4). The images in Figures 1, 2, and 3D were taken using a 100× oil immersion objective with a phase ring (NA 1.45, Ph 3, Nikon). For wide-field fluorescence imaging, the sample was illuminated with monochromatic LED light of 640 nm (Lumencor Spectra Pad X). Confocal images were gathered on the same imaging setup using a spinning disk confocal (Crest X-light) with pinhole size 70 μm. Phase contrast images were acquired by illuminating with the microscope’s DIA illuminator and using the phase mask in the microscope’s condenser. For the image shown in Figure 3A, the coverslip with PVA gel and dried lipids was put on the microscope. The image was taken in wide-field fluorescence mode using a 60× long working distance water immersion objective (CFI Plan Apochromat VS 60× WI, Nikon) 10 min after the addition of the swelling buffer.
Actin-deformed GUVs shown in Figures 3C and 5E were produced by F.C. Tsai as described in detail in ref (40). In short, GUVs of a lipid composition of DOPC:Rhodamine-DOPE:PEG2000DPPE in a molar ratio of 94.8:0.2:5 were produced by gel-assisted swelling on top of an agarose gel. Actin was encapsulated by adding it to the swelling solution at a concentration of 12 μM, and at a 5:1 molar ratio with respect to fascin. 20–30 mol % of the actin was labeled with Alexa 488 to allow fluorescence visualization. After formation, GUVs were harvested and imaged by confocal fluorescence microscopy. Images were taken with a Nikon Eclipse Ti inverted microscope equipped with a Nikon C1 confocal scanhead, a 100× NA 1.4 Plan Apo oil immersion objective and lasers with wavelengths 488 and 543 nm.
Microfluidic vesicle production (Figure 3B) was done with the octanol-assisted liposome assembly (OLA) technique following ref (22). Lipids were used in a composition of DOPC: Rhodamine-DOPE in a molar ratio 99.5:0.5. The inner aqueous solution consisted of 5% (v/v) glycerol in Milli-Q water, and the outer solution of 15% (v/v) glycerol and 5% (w/v) Poloxamer 188. GUVs were imaged directly on-chip in the postformation channel with a 10× air objective (Plan Fluor, NA 0.3, Nikon).
Phase-separated GUVs (Figure 5) were produced by gel-assisted swelling as described above, but with minor modifications. Lipids were dried in a mixture of DOPC:DPPC:cholesterol:NBD-DPPE:ATTO655-DOPE in molar ratio 31.8:48:20:0.1:0.1. In addition, swelling was done in a 37 °C room to be above the membrane transition temperature (around 35.5 °C98), and thus to ensure proper mixing of lipids during formation.
4.1.3. GUVs for Population Analysis
DNA-mediated vesicle binding was performed following refs (90) and (91). GUVs with a membrane composed of DOPC:ATTO 488 DOPE in molar ratio 99.5:0.5 were produced by gel-assisted swelling as described above in a solution containing 100 mOsm sucrose, 100 mM KCl and 10 mM Tris-HCl at pH 7.4. LUVs were produced by adding lipids in chloroform to a Pyrex glass tube, in a lipid composition of DOPC:ATTO 655 DOPE as 99.95:0.05 (mol/mol). After drying lipids for 1 h in a vacuum desiccator, the dried film was resuspended by vortexing for 2 min in a solution containing 100 mM KCl and 10 mM Tris-HCl at pH 7.4 to a final lipid concentration of 0.5 mg/mL. To produce 200 nm LUVs, the suspension was extruded (Mini Extruder, Avanti Polar Lipids, Inc.) 21 times over a polycarbonate membrane with pore size 200 nm (Nuclepore, Whatman). To introduce specific binding between LUVs and GUVs, we used two complementary DNA strands (DNA1 and DNA1′) with a cholesterol moiety for membrane anchoring90 (biomers.net, Ulm, Germany). The DNA strands were tagged with cholesterol on opposite ends to allow antiparallel binding, as is typically used for DNA-mediated membrane fusion assays.99 The full sequences were taken from ref (91) and read:
chol-DNA1: 5′-TGGACATCAGAAAGGCACGACGA-cholesterol-TEG-3′
chol-DNA1′: cholesterol-TEG-5′-TCCGTCGTGCCTTATTTCTGATGTCCA-3′
Note that the sequences do not fully overlap, which results from an error in the original publication (Y. Dreher, personal communication, 2021). For 1 h, LUVs were incubated with 1 μM chol-DNA1, and GUVs with 1 μM chol-DNA1′. After DNA incubation, GUVs and LUVs were mixed, left to bind for 1 h, and finally imaged in a solution containing 100 mOsm glucose, 100 mM KCl, and 10 mM Tris-HCl at pH 7.4. Images were taken in wide-field fluorescence mode using a 100× oil immersion objective (CFI Plan Apochromat VC 100× oil, NA 1.40, Nikon) at 508 and 640 nm to image LUVs and GUVs, respectively.
Vesicles containing monomeric actin were produced as in ref (25). Lipids were mixed in a DOPC:PEG2000-DOPE:Cy5-DOPE molar ratio of 99.89:0.01:0.1. We encapsulated 4.4 μM actin in G-buffer, of which 10% was labeled with Alexa-488 to allow fluorescence visualization. The encapsulated solution also included 1 mM dithiothreitol (DTT), 1 μM protocatechuic acid (PCA), and 1 μM protocatechuate dioxygenase (PCD). Wide-field fluorescence images were taken with a 100× oil immersion objective (CFI Plan Apochromat VC 100× oil, NA 1.40, Nikon) at a wavelength of 640 and 470 nm to visualize the vesicle membrane and actin content, respectively.
4.2. Analysis
4.2.1. Vesicle Detection
Raw images containing vesicles are loaded in the software and can be preprocessed in two steps: smoothing and membrane enhancement. Image smoothing is performed by convolution of the original image with a two-dimensional (2D) Gaussian function of user-defined kernel size. Membrane enhancement is based on the subtraction of a second smoothed image (the subtraction image) from the first smoothed image to eliminate large-scale intensity variations. To this end, the user specifies a kernel size (typically 10–20 times larger than the kernel used for smoothing the image) to create the subtraction image. The subtraction image is then subtracted from the smoothed image to create the enhanced image.
Vesicles were detected in the images with one of DisGUVery’s detection methods: circular Hough transform (CHT), multiscale template matching (MTM), and floodfill detection (FF). CHT is an established algorithm for identifying circular objects.63,100 MTM is a template matching algorithm in which we have added a template rescaling option to enable size-invariant detection. Vesicle locations correspond to points with a high correlation coefficient between the image and the used template. Vesicles detected at different scales are sorted by their correlation coefficient to keep only the scale with the highest template matching score. Detection by FF starts with a binarization of the image based on absolute intensity, followed by flooding the background pixels in the extracellular solution. All methods were implemented using the package OpenCV(101) and the corresponding functions as a base. After detection, falsely detected vesicles can be discarded.
To evaluate the performance of vesicle detection, we used three data sets of different imaging types: confocal fluorescence, wide-field fluorescence, and phase contrast. Each set consisted of five images. Using DisGUVery, images were preprocessed by smoothing and enhancement as described above, and vesicles were detected with one of the three algorithms. We used detection input settings that resulted in both high precision and high recall as was verified by visual inspection. Detection results were then compared with manual evaluation to determine precision and recall and from there we calculated the F1 score (see main text). To map the dependence of the F1 score on input settings, we systematically varied two settings and evaluated detection in the five confocal images for each unique combination. Vesicle classification was performed by visual inspection by four testers in five confocal images. Counts of vesicles in the different categories are shown in Table S1.
4.2.2. Membrane Segmentation
After detection, DisGUVery can be used to segment the membrane area of vesicles in one of two ways: directly from the vesicle detection output, called basic membrane analysis (BMA), or with an extra contour tracking step, called refined membrane detection (RMD). In BMA, a circular ring-shaped region of interest (ROI) that surrounds the membrane is created based on the vesicle detection result.
Membrane tracking with RMD consists of a modified Canny edge detection algorithm102,103 combined with a directional search algorithm that we developed to chain edge points and to assign them to either the inner or the outer edge of the membrane. The search occurs at each detected edge point, chosen at random, by chaining the neighboring points contained within a defined search box centered around the point of interest and typically of aspect ratio > 2:1 (length: width). The orientation of the box is determined as orthogonal to the direction of the steepest gradient (across the membrane). Finally, all chains are measured by the number of points within, and the two longest chains will be assigned to the outer and inner edge of the membrane (Figure 5B). This directional search with a bounding box allows us to chain points together without the need of them being connected. Furthermore, it enables the user to distinguish the enclosing GUV membrane, which is the membrane separating the inner from the outer solution, from internal membranes and secondary membrane structures such as tubes. Dependent on the membrane appearance, for example, its thickness in the image or the presence of secondary membrane structures, the user can define the length and width of the search box to fine-tune the tracking results. By calculating the center of mass of the detected contours, the vesicle center is refined after RMD.
4.2.3. Analysis Modules
DisGUVery enables the user to use a set of predefined analysis modules, including vesicle size distributions, angular and radial intensity profiles, and calculation of internal fluorescence.
The intensity profiles used here (Figures 5 and 6A–E) were obtained in DisGUVery. After vesicle detection, membranes were segmented with BMA (Figures 5F and 6E) or RMD (Figure 5E). From the membrane segmentation, angular and radial intensity profiles were computed in the software. In addition, angular distances were obtained from the RMD contour (Figure 5E). The angular intensity profiles and angular distances were exported in a results file and further postprocessing (normalization, plotting) was done with a custom-written Python script outside the software. For Figure 6E, postprocessing included determination of the median intensity, background subtraction, statistical tests, and producing the bar plot.
Encapsulation analysis was done by first detecting vesicles with FF and directly using the FF masks in DisGUVery to calculate the internal vesicle fluorescence from the vesicle lumen in both color channels. Postprocessing (background subtraction, normalization, linear regression, plotting) was done outside the software with a custom-written script.
Acknowledgments
The authors thank Martin Depken, Rumiana Dimova, and Carlos Marques for useful discussions; Britta Bor, Gerard Castro Linares, Lucia Baldauf, and Nikki Nafar for performance evaluation of vesicle detection; Tom Aarts and Lucia Baldauf for images on DNA-mediated vesicle binding; Jeffrey den Haan for protein purification and labeling; and the Biological Soft Matter group for testing and feedback. They acknowledge the ‘BaSyC - Building a Synthetic Cell’ Gravitation grant of the Netherlands Ministry of Education, Culture and Science (OCW) and the Netherlands Organization for Scientific Research (NWO) for funding.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.2c00407.
Author Present Address
‡ Institute of Physics and Astronomy, University of Potsdam, 14476 Potsdam, Germany
Author Contributions
All authors conceived the idea and designed the project. L.v.B. performed the experiments. C.M.T. developed the software. L.v.B. performed testing and performance evaluation. L.v.B. and C.M.T. performed analysis of the results and wrote the first draft of the manuscript. All authors contributed to manuscript revision and read and approved the submitted version.
The authors declare no competing financial interest.
Supplementary Material
References
- Walde P.; Cosentino K.; Engel H.; Stano P. Giant Vesicles: Preparations and Applications. ChemBioChem 2010, 11, 848–865. 10.1002/cbic.201000010. [DOI] [PubMed] [Google Scholar]
- Henriksen J.; Rowat A. C.; Ipsen J. H. Vesicle fluctuation analysis of the effects of sterols on membrane bending rigidity. Eur. Biophys. J. 2004, 33, 732–741. 10.1007/s00249-004-0420-5. [DOI] [PubMed] [Google Scholar]
- Ruan Q.; Cheng M. A.; Levi M.; Gratton E.; Mantulin W. W. Spatial-Temporal Studies of Membrane Dynamics: Scanning Fluorescence Correlation Spectroscopy (SFCS). Biophys. J. 2004, 87, 1260–1267. 10.1529/biophysj.103.036483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamba Y.; Yamazak M. Single Giant Unilamellar Vesicle Method Reveals Effect of Antimicrobial Peptide Magainin 2 on Membrane Permeability. Biochemistry 2005, 44, 15823–15833. 10.1021/bi051684w. [DOI] [PubMed] [Google Scholar]
- Kaiser H.-J.; Lingwood D.; Levental I.; Sampaio J. L.; Kalvodova L.; Rajendran L.; Simons K. Order of lipid phases in model and plasma membranes. Proc. Natl. Acad. Sci. 2009, 106, 16645–16650. 10.1073/pnas.0908987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagatolli L. A.; Gratton E. A Correlation between Lipid Domain Shape and Binary Phospholipid Mixture Composition in Free Standing Bilayers: A Two-Photon Fluorescence Microscopy Study. Biophys. J. 2000, 79, 434–447. 10.1016/S0006-3495(00)76305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicuta P.; Keller S. L.; Veatch S. L. Diffusion of Liquid Domains in Lipid Bilayer Membranes. J. Phys. Chem. B 2007, 111, 3328–3331. 10.1021/jp0702088. [DOI] [PubMed] [Google Scholar]
- Blanken D.; Foschepoth D.; Serrão A. C.; Danelon C. Genetically controlled membrane synthesis in liposomes. Nat. Commun. 2020, 11, 4317 10.1038/s41467-020-17863-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkühler J.; Knorr R. L.; Zhao Z.; Bhatia T.; Bartelt S. M.; Wegner S.; Dimova R.; Lipowsky R. Controlled division of cell-sized vesicles by low densities of membrane-bound proteins. Nat. Commun. 2020, 11, 905 10.1038/s41467-020-14696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T.; Wunder C.; Lippincott-Schwartz J.; Hurley J. H. Membrane scission by the ESCRT-III complex. Nature 2009, 458, 172–177. 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T.; Yamazaki M. Membrane Fusion of Giant Unilamellar Vesicles of Neutral Phospholipid Membranes Induced by La3+. Langmuir 2004, 20, 5160–5164. 10.1021/la049681s. [DOI] [PubMed] [Google Scholar]
- Mulla Y.; Aufderhorst-Roberts A.; Koenderink G. H. Shaping up synthetic cells. Phys. Biol. 2018, 15, 041001 10.1088/1478-3975/aab923. [DOI] [PubMed] [Google Scholar]
- Abil Z.; Danelon C. Roadmap to Building a Cell: An Evolutionary Approach. Front. Bioeng. Biotechnol. 2020, 8, 927 10.3389/fbioe.2020.00927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A. P.; Fletcher D. A. Biology under construction: in vitro reconstitution of cellular function. Nat. Rev. Mol. Cell Biol. 2009, 10, 644–650. 10.1038/nrm2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwille P. Bottom-up synthetic biology: Engineering in a Tinkerer’s World. Science 2011, 333, 1252–1254. 10.1126/science.1211701. [DOI] [PubMed] [Google Scholar]
- Lussier F.; Staufer O.; Platzman I.; Spatz J. P. Can Bottom-Up Synthetic Biology Generate Advanced Drug-Delivery Systems?. Trends Biotechnol. 2021, 39, 445–459. 10.1016/j.tibtech.2020.08.002. [DOI] [PubMed] [Google Scholar]
- Angelova M. I.; Dimitrov D. S. Liposome electroformation. Faraday Discuss. Chem. Soc. 1986, 81, 303–311. 10.1039/dc9868100303. [DOI] [Google Scholar]
- Nourian Z.; Roelofsen W.; Danelon C. Triggered Gene Expression in Fed-Vesicle Microreactors with a Multifunctional Membrane. Angew. Chem., Int. Ed. 2012, 51, 3114–3118. 10.1002/anie.201107123. [DOI] [PubMed] [Google Scholar]
- Weinberger A.; Tsai F.-C.; Koenderink G. H.; Schmidt T.; Itri R.; Meier W.; Scmatko T.; Schröder A.; Marques C. Gel-assisted formation of giant unilamellar vesicles. Biophys. J. 2013, 105, 154–164. 10.1016/j.bpj.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautot S.; Frisken B. J.; Weitz D. A. Production of Unilamellar Vesicles Using an Inverted Emulsion. Langmuir 2003, 19, 2870–2879. 10.1021/la026100v. [DOI] [Google Scholar]
- Moga A.; Yandrapalli D. N.; Dimova D. R.; Robinson D. T.; Yandrapalli N.; Dimova R.; Robinson T. Optimization of the Inverted Emulsion Method for High-Yield Production of Biomimetic Giant Unilamellar Vesicles. ChemBioChem 2019, 20, 2674 10.1002/cbic.201900529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande S.; Caspi Y.; Meijering A.; Dekker C. Octanol-assisted liposome assembly on chip. Nat. Commun. 2016, 7, 10447 10.1038/ncomms10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng N.-N.; Vibhute M. A.; Zheng L.; Zhao H.; Yelleswarapu M.; Huck W. T. S. Macromolecularly Crowded Protocells from Reversibly Shrinking Monodisperse Liposomes. J. Am. Chem. Soc. 2018, 140, 7399–7402. 10.1021/jacs.8b03123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abkarian M.; Loiseau E.; Massiera G. Continuous droplet interface crossing encapsulation (cDICE) for high throughput monodisperse vesicle design. Soft Matter 2011, 7, 4610–4614. 10.1039/c1sm05239j. [DOI] [Google Scholar]
- Van de Cauter L.; Fanalista F.; van Buren L.; Franceschi N. D.; Godino E.; Bouw S.; Danelon C.; Dekker C.; Koenderink G. H.; Ganzinger K. A. Optimized cDICE for Efficient Reconstitution of Biological Systems in Giant Unilamellar Vesicles. ACS Synth. Biol. 2021, 10, 1690–1702. 10.1021/acssynbio.1c00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes L. R.; Alonso A.; Goñi F. M.; Bagatolli L. A. Giant Unilamellar Vesicles Electroformed from Native Membranes and Organic Lipid Mixtures under Physiological Conditions. Biophys. J. 2007, 93, 3548–3554. 10.1529/biophysj.107.116228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maktabi S.; Schertzer J. W.; Chiarot P. R. Dewetting-induced formation and mechanical properties of synthetic bacterial outer membrane models (GUVs) with controlled inner-leaflet lipid composition. Soft Matter 2019, 15, 3938–3948. 10.1039/C9SM00223E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira R. B.; Robinson T.; Dimova R.; Riske K. A. Highly Efficient Protein-free Membrane Fusion: A Giant Vesicle Study. Biophys. J. 2019, 116, 79–91. 10.1016/j.bpj.2018.11.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blosser M. C.; Horst B. G.; Keller S. L. cDICE method produces giant lipid vesicles under physiological conditions of charged lipids and ionic solutions. Soft Matter 2016, 12, 7364–7371. 10.1039/C6SM00868B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürre K.; Keber F. C.; Bleicher P.; Brauns F.; Cyron C. J.; Faix J.; Bausch A. R. Capping protein-controlled actin polymerization shapes lipid membranes. Nat. Commun. 2018, 9, 1630 10.1038/s41467-018-03918-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamdad K.; Law R. V.; Seddon J. M.; Brooks N. J.; Ces O. Studying the effects of asymmetry on the bending rigidity of lipid membranes formed by microfluidics. Chem. Commun. 2016, 52, 5277–5280. 10.1039/C5CC10307J. [DOI] [PubMed] [Google Scholar]
- Girard P.; Pécréaux J.; Lenoir G.; Falson P.; Rigaud J. L.; Bassereau P. A New Method for the Reconstitution of Membrane Proteins into Giant Unilamellar Vesicles. Biophys. J. 2004, 87, 419–429. 10.1529/biophysj.104.040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott T.; Bouvrais H.; Méléard P. Giant unilamellar vesicle formation under physiologically relevant conditions. Chem. Phys. Lipids 2008, 154, 115–119. 10.1016/j.chemphyslip.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Keber F. C.; Loiseau E.; Sanchez T.; DeCamp S. J.; Giomi L.; Bowick M. J.; Marchetti M. C.; Dogic Z.; Bausch A. R. Topology and dynamics of active nematic vesicles. Science 2014, 345, 1135–1139. 10.1126/science.1254784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vutukuri H. R.; Hoore M.; Abaurrea-Velasco C.; van Buren L.; Dutto A.; Auth T.; Fedosov D. A.; Gompper G.; Vermant J. Active particles induce large shape deformations in giant lipid vesicles. Nature 2020, 586, 52–56. 10.1038/s41586-020-2730-x. [DOI] [PubMed] [Google Scholar]
- García-Calvo J.; Maillard J.; Fureraj I.; Strakova K.; Colom A.; Mercier V.; Roux A.; Vauthey E.; Sakai N.; Fürstenberg A.; Matile S. Fluorescent Membrane Tension Probes for Super-Resolution Microscopy: Combining Mechanosensitive Cascade Switching with Dynamic-Covalent Ketone Chemistry. J. Am. Chem. Soc. 2020, 142, 12034–12038. 10.1021/jacs.0c04942. [DOI] [PubMed] [Google Scholar]
- Sato K.; Obinata K.; Sugawara T.; Urabe I.; Yomo T. Quantification of structural properties of cell-sized individual liposomes by flow cytometry. J. Biosci. Bioeng. 2006, 102, 171–178. 10.1263/jbb.102.171. [DOI] [PubMed] [Google Scholar]
- Boban Z.; Mardeši’c I. M.; Subczynski W. K.; Raguz M.; Koty’nska J.; Koty’nska K.; Naumowicz M. Giant Unilamellar Vesicle Electroformation: What to Use, What to Avoid, and How to Quantify the Results. Membranes 2021, 11, 860 10.3390/membranes11110860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba M.; Miyazaki M.; Ishiwata S. Quantitative Analysis of the Lamellarity of Giant Liposomes Prepared by the Inverted Emulsion Method. Biophys. J. 2014, 107, 346–354. 10.1016/j.bpj.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai F.-C.; Koenderink G. Shape control of lipid bilayer membranes by confined actin bundles. Soft Matter 2015, 11, 8834–8847. 10.1039/C5SM01583A. [DOI] [PubMed] [Google Scholar]
- Li Q.; Wang X.; Ma S.; Zhang Y.; Han X. Electroformation of giant unilamellar vesicles in saline solution. Colloids Surf., B 2016, 147, 368–375. 10.1016/j.colsurfb.2016.08.018. [DOI] [PubMed] [Google Scholar]
- Deshpande S.; Wunnava S.; Hueting D.; Dekker C. Membrane Tension-Mediated Growth of Liposomes. Small 2019, 15, 1902898 10.1002/smll.201902898. [DOI] [PubMed] [Google Scholar]
- Parigoris E.; Dunkelmann D. L.; Murphy A.; Wili N.; Kaech A.; Dumrese C.; Jimenez-Rojo N.; Silvan U. Facile generation of giant unilamellar vesicles using polyacrylamide gels. Sci. Rep. 2020, 10, 8461 10.1038/s41598-020-61655-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wubshet N. H.; Bashirzadeh Y.; Liu A. P. Fascin-induced actin protrusions are suppressed by dendritic networks in giant unilamellar vesicles. Mol. Biol. Cell 2021, 32, 1634–1640. 10.1091/mbc.E21-02-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudheti M. V.; Mlodzianoski M.; Hess S. T. Imaging and Shape Analysis of GUVs as Model Plasma Membranes: Effect of Trans DOPC on Membrane Properties. Biophys. J. 2007, 93, 2011–2023. 10.1529/biophysj.106.103374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanken D.; Van Nies P.; Danelon C. Quantitative imaging of gene-expressing liposomes reveals rare favorable phenotypes. Phys. Biol. 2019, 16, 045002 10.1088/1478-3975/ab0c62. [DOI] [PubMed] [Google Scholar]
- Staufer O.; Antona S.; Zhang D.; Csatári J.; Schröter M.; Janiesch J. W.; Fabritz S.; Berger I.; Platzman I.; Spatz J. P. Microfluidic production and characterization of biofunctionalized giant unilamellar vesicles for targeted intracellular cargo delivery. Biomaterials 2021, 264, 120203 10.1016/j.biomaterials.2020.120203. [DOI] [PubMed] [Google Scholar]
- Guiet R.; Burri O.; Seitz A. Open source tools for biological image analysis. Methods Mol. Biol. 2019, 2040, 23–37. 10.1007/978-1-4939-9686-5_2. [DOI] [PubMed] [Google Scholar]
- Hermann E.; Bleicken S.; Subburaj Y.; García-Sáez A. J. Automated analysis of giant unilamellar vesicles using circular Hough transformation. Bioinformatics 2014, 30, 1747–1754. 10.1093/bioinformatics/btu102. [DOI] [PubMed] [Google Scholar]
- Sych T.; Schubert T.; Vauchelles R.; Madl J.; Omidvar R.; Thuenauer R.; Richert L.; Mély Y.; Römer W. GUV-AP: multifunctional FIJI-based tool for quantitative image analysis of Giant Unilamellar Vesicles. Bioinformatics 2019, 35, 2340–2342. 10.1093/bioinformatics/bty962. [DOI] [PubMed] [Google Scholar]
- Leomil F. S. C.; Zoccoler M.; Dimova R.; Riske K. A.; Bateman A. PoET: automated approach for measuring pore edge tension in giant unilamellar vesicles. Bioinform. Adv. 2021, 1, vbab037 10.1093/bioadv/vbab037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husen P.; Arriaga L. R.; Monroy F.; Ipsen J. H.; Bagatolli L. A. Morphometric Image Analysis of Giant Vesicles: A New Tool for Quantitative Thermodynamics Studies of Phase Separation in Lipid Membranes. Biophys. J. 2012, 103, 2304–2310. 10.1016/j.bpj.2012.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.-H.; Passaro S.; Ozturk S.; Wang W.. Intelligent Fluorescence Image Analysis of Giant Unilamellar Vesicles and Protein Liquid Phase Droplets by Whole Z-stack Analysis; Research Square, 2021. [Google Scholar]
- Beber A.; Alqabandi M.; Prévost C.; Viars F.; Lévy D.; Bassereau P.; Bertin A.; Mangenot S. Septin-based readout of PI(4,5)P2 incorporation into membranes of giant unilamellar vesicles. Cytoskeleton 2019, 76, 92–103. 10.1002/cm.21480. [DOI] [PubMed] [Google Scholar]
- Barooji Y. F.; Rørvig-Lund A.; Semsey S.; Reihani S. N. S.; Bendix P. M. Dynamics of membrane nanotubes coated with I-BAR. Sci. Rep. 2016, 6, 30054 10.1038/srep30054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental K. R.; Levental I. Giant Plasma Membrane Vesicles: Models for Understanding Membrane Organization. Curr. Top. Membr. 2015, 75, 25–57. 10.1016/bs.ctm.2015.03.009. [DOI] [PubMed] [Google Scholar]
- Loiseau E.; Schneider J. A.; Keber F. C.; Pelzl C.; Massiera G.; Salbreux G.; Bausch A. R. Shape remodeling and blebbing of active cytoskeletal vesicles. Sci. Adv. 2016, 2, e1500465 10.1126/sciadv.1500465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandrapalli N.; Petit J.; Bäumchen O.; Robinson T. Surfactant-free production of biomimetic giant unilamellar vesicles using PDMS-based microfluidics. Commun. Chem. 2021, 4, 1–10. 10.1038/s42004-021-00530-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göpfrich K.; Haller B.; Staufer O.; Dreher Y.; Mersdorf U.; Platzman I.; Spatz J. P. One-Pot Assembly of Complex Giant Unilamellar Vesicle-Based Synthetic Cells. ACS Synth. Biol. 2019, 8, 937–947. 10.1021/acssynbio.9b00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudji I. R.; Subburaj Y.; Frenkel N.; García-Sáez A. J.; Wink M. Membrane Disintegration Caused by the Steroid Saponin Digitonin Is Related to the Presence of Cholesterol. Molecules 2015, 20, 20146–20160. 10.3390/molecules201119682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei B.; Lira R. B.; Perez K. R.; Riske K. A. Membrane permeabilization induced by Triton X-100: The role of membrane phase state and edge tension. Chem. Phys. Lipids 2017, 202, 28–37. 10.1016/j.chemphyslip.2016.11.009. [DOI] [PubMed] [Google Scholar]
- Nishimura K.; Matsuura T.; Nishimura K.; Sunami T.; Suzuki H.; Yomo T. Cell-free protein synthesis inside giant unilamellar vesicles analyzed by flow cytometry. Langmuir 2012, 28, 8426–8432. 10.1021/la3001703. [DOI] [PubMed] [Google Scholar]
- Duda R. O.; Hart P. E. Use of the Hough transformation to detect lines and curves in pictures. Commun. ACM 1972, 15, 11–15. 10.1145/361237.361242. [DOI] [Google Scholar]
- Brunelli R.Template Matching Techniques in Computer Vision: Theory and Practice; John Wiley and Sons, Ltd, 2009. [Google Scholar]
- Foley J. D.; Van Dam A.. Fundamentals of Interactive Computer Graphics; Addison-Wesley Longman Publishing Co., Inc., 1982. [Google Scholar]
- Chinchor N.; Chinchor N. In MUC-4 Evaluation Metrics, 4th Message Understanding Conference, MUC, 1992; pp 22–29.
- Chicco D.; Jurman G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics 2020, 21, 0208737 10.1186/s12864-019-6413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideau E.; Wurm F. R.; Landfester K. Self-Assembly of Giant Unilamellar Vesicles by Film Hydration Methodologies. Adv. Biosyst. 2019, 3, 1800324 10.1002/adbi.201800324. [DOI] [PubMed] [Google Scholar]
- Souissi M.; Pernier J.; Rossier O.; Giannone G.; Le Clainche C.; Helfer E.; Sengupta K. Integrin-Functionalised Giant Unilamellar Vesicles via Gel-Assisted Formation: Good Practices and Pitfalls. Int. J. Mol. Sci. 2021, 22, 6335 10.3390/ijms22126335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartelt S. M.; Chervyachkova E.; Ricken J.; Wegner S. V. Mimicking Adhesion in Minimal Synthetic Cells. Adv. Biosyst. 2019, 3, 1800333 10.1002/adbi.201800333. [DOI] [PubMed] [Google Scholar]
- Wang X.; Tian L.; Du H.; Li M.; Mu W.; Drinkwater B. W.; Han X.; Mann S. Chemical communication in spatially organized protocell colonies and protocell/living cell micro-arrays. Chem. Sci. 2019, 10, 9446–9453. 10.1039/C9SC04522H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supramaniam P.; Ces O.; Salehi-Reyhani A. Microfluidics for Artificial Life: Techniques for Bottom-Up Synthetic Biology. Micromachines 2019, 10, 299 10.3390/mi10050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai Y.; Xie R.; Xiong J.; Liang Q. Microfluidics for Biosynthesizing: from Droplets and Vesicles to Artificial Cells. Small 2020, 16, 1903940 10.1002/smll.201903940. [DOI] [PubMed] [Google Scholar]
- Litschel T.; Ramm B.; Maas R.; Heymann M.; Schwille P. Beating Vesicles: Encapsulated Protein Oscillations Cause Dynamic Membrane Deformations. Angew. Chem., Int. Ed. 2018, 57, 16286–16290. 10.1002/anie.201808750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganzinger K. A.; Merino-Salomón A.; García-Soriano D. A.; Butterfield A. N.; Litschel T.; Siedler F.; Schwille P. FtsZ Reorganization Facilitates Deformation of Giant Vesicles in Microfluidic Traps. Angew. Chem. 2020, 132, 21556–21560. 10.1002/ange.202001928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng N. N.; Yelleswarapu M.; Huck W. T. Monodisperse Uni- and Multicompartment Liposomes. J. Am. Chem. Soc. 2016, 138, 7584–7591. 10.1021/jacs.6b02107. [DOI] [PubMed] [Google Scholar]
- Sobrinos-Sanguino M.; Zorrilla S.; Keating C. D.; Monterroso B.; Rivas G. Encapsulation of a compartmentalized cytoplasm mimic within a lipid membrane by microfluidics. Chem. Commun. 2017, 53, 4775–4778. 10.1039/C7CC01289F. [DOI] [PubMed] [Google Scholar]
- Martinez-Torres C. E.; Laperrousaz B.; Berguiga L.; Boyer-Provera E.; Elezgaray J.; MD F. E. N.; Maguer-Satta V.; Arneodo A.; Argoul J. Deciphering the internal complexity of living cells with quantitative phase microscopy: a multiscale approach. J. Biomed. Opt. 2015, 20, 096005 10.1117/1.JBO.20.9.096005. [DOI] [PubMed] [Google Scholar]
- Bashirzadeh Y.; Wubshet N. H.; Liu A. P. Confinement Geometry Tunes Fascin-Actin Bundle Structures and Consequently the Shape of a Lipid Bilayer Vesicle. Front. Mol. Biosci. 2020, 7, 610277 10.3389/fmolb.2020.610277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirzadeh Y.; Moghimianavval H.; Liu A. Encapsulated Actomyosin Patterns Drive Cell-Like Membrane Shape Changes. SSRN Electron. J. 2021, 25, 104236 10.2139/ssrn.3956818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litschel T.; Kelley C. F.; Holz D.; Adeli Koudehi M.; Vogel S. K.; Burbaum L.; Mizuno N.; Vavylonis D.; Schwille P. Reconstitution of contractile actomyosin rings in vesicles. Nat. Commun. 2021, 12, 2254 10.1038/s41467-021-22422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Diaz D. A.; Merino-Salomón A.; Meyer F.; Heymann M.; Rivas G.; Bramkamp M.; Schwille P. FtsZ induces membrane deformations via torsional stress upon GTP hydrolysis. Nat. Commun. 2021, 12, 3310 10.1038/s41467-021-23387-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czogalla A.; Kauert D. J.; Franquelim H. G.; Uzunova V.; Zhang Y.; Seidel R.; Schwille P. Amphipathic DNA Origami Nanoparticles to Scaffold and Deform Lipid Membrane Vesicles. Angew. Chem., Int. Ed. 2015, 54, 6501–6505. 10.1002/anie.201501173. [DOI] [PubMed] [Google Scholar]
- Fanalista F.; Birnie A.; Maan R.; Burla F.; Charles K.; Pawlik G.; Deshpande S.; Koenderink G. H.; Dogterom M.; Dekker C. Shape and Size Control of Artificial Cells for Bottom-Up Biology. ACS Nano 2019, 13, 5439–5450. 10.1021/acsnano.9b00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pécréaux J.; Döbereiner H. G.; Prost J.; Joanny J. F.; Bassereau P. Refined contour analysis of giant unilamellar vesicles. Eur. Phys. J. E 2004, 13, 277–290. 10.1140/epje/i2004-10001-9. [DOI] [PubMed] [Google Scholar]
- Liu A. P.; Richmond D. L.; Maibaum L.; Pronk S.; Geissler P. L.; Fletcher D. A. Membrane-induced bundling of actin filaments. Nat. Phys. 2008, 4, 789–793. 10.1038/nphys1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veatch S. L.; Keller S. L. Separation of Liquid Phases in Giant Vesicles of Ternary Mixtures of Phospholipids and Cholesterol. Biophys. J. 2003, 85, 3074–3083. 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkühler J.; De Tillieux P.; Knorr R. L.; Lipowsky R.; Dimova R. Charged giant unilamellar vesicles prepared by electroformation exhibit nanotubes and transbilayer lipid asymmetry. Sci. Rep. 2018, 8, 11838 10.1038/s41598-018-30286-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro M.; Reina F.; Hof M.; Eggeling C.; Sezgin E. Laurdan and Di-4-ANEPPDHQ probe different properties of the membrane. J. Phys. D: Appl. Phys. 2017, 50, 134004 10.1088/1361-6463/aa5dbc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel G.; Zahn R.; Höök F. DNA-induced programmable fusion of phospholipid vesicles. J. Am. Chem. Soc. 2007, 129, 9584–9585. 10.1021/ja073200k. [DOI] [PubMed] [Google Scholar]
- Dreher Y.; Jahnke K.; Bobkova E.; Spatz J. P.; Göpfrich K. Division and Regrowth of Phase-Separated Giant Unilamellar Vesicles. Angew. Chem. 2021, 133, 10756–10764. 10.1002/ange.202014174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivi L.; Berger M.; Creyghton R. N.; De Franceschi N.; Dekker C.; Mulder B. M.; Claassens N. J.; ten Wolde P. R.; van der Oost J. Towards a synthetic cell cycle. Nat. Commun. 2021, 12, 4531 10.1038/s41467-021-24772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litschel T.; Schwille P. Protein Reconstitution Inside Giant Unilamellar Vesicles. Annu. Rev. Biophys. 2021, 50, 525–548. 10.1146/annurev-biophys-100620-114132. [DOI] [PubMed] [Google Scholar]
- Ganzinger K. A.; Schwille P. More from less—Bottom-up reconstitution of cell biology. J. Cell Sci. 2019, 132, jcs227488 10.1242/jcs.227488. [DOI] [PubMed] [Google Scholar]
- Pratapa A.; Doron M.; Caicedo J. C. Image-based cell phenotyping with deep learning. Curr. Opin. Chem. Biol. 2021, 65, 9–17. 10.1016/j.cbpa.2021.04.001. [DOI] [PubMed] [Google Scholar]
- Alvarado J.; Koenderink G. H.. Reconstituting Cytoskeletal Contraction Events with Biomimetic Actin-Myosin Active Gels. In Methods in Cell Biology; Academic Press, 2015; Vol. 128, pp 83–103. [DOI] [PubMed] [Google Scholar]
- Gentry B. S.; Van Der Meulen S.; Noguera P.; Alonso-Latorre B.; Plastino J.; Koenderink G. H. Multiple actin binding domains of Ena/VASP proteins determine actin network stiffening. Eur. Biophys. J. 2012, 41, 979–990. 10.1007/s00249-012-0861-1. [DOI] [PubMed] [Google Scholar]
- Juhasz J.; Sharom F. J.; Davis J. H. Quantitative characterization of coexisting phases in DOPC/DPPC/cholesterol mixtures: Comparing confocal fluorescence microscopy and deuterium nuclear magnetic resonance. Biochim. Biophys. Acta, Biomembr. 2009, 1788, 2541–2552. 10.1016/j.bbamem.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Chan Y.-H. M.; van Lengerich B.; Boxer S. G. Lipid-anchored DNA mediates vesicle fusion as observed by lipid and content mixing. Biointerphases 2008, 3, FA17–FA21. 10.1116/1.2889062. [DOI] [PubMed] [Google Scholar]
- Kimme C.; Ballard D.; Sklansky J. Finding circles by an array of accumulators. Commun. ACM 1975, 18, 120–122. 10.1145/360666.360677. [DOI] [Google Scholar]
- Bradski G.The OpenCV Library, Dr. Dobb’s Journal of Software Tools, M&T Publication, 2000.
- Canny J. A Computational Approach to Edge Detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, 679–698. 10.1109/TPAMI.1986.4767851. [DOI] [PubMed] [Google Scholar]
- Martinez-Torres C.; Burla F.; Alkemade C.; Koenderink G. H. Revealing the assembly of filamentous proteins with scanning transmission electron microscopy. PLoS One 2019, 14, e0226277 10.1371/journal.pone.0226277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.