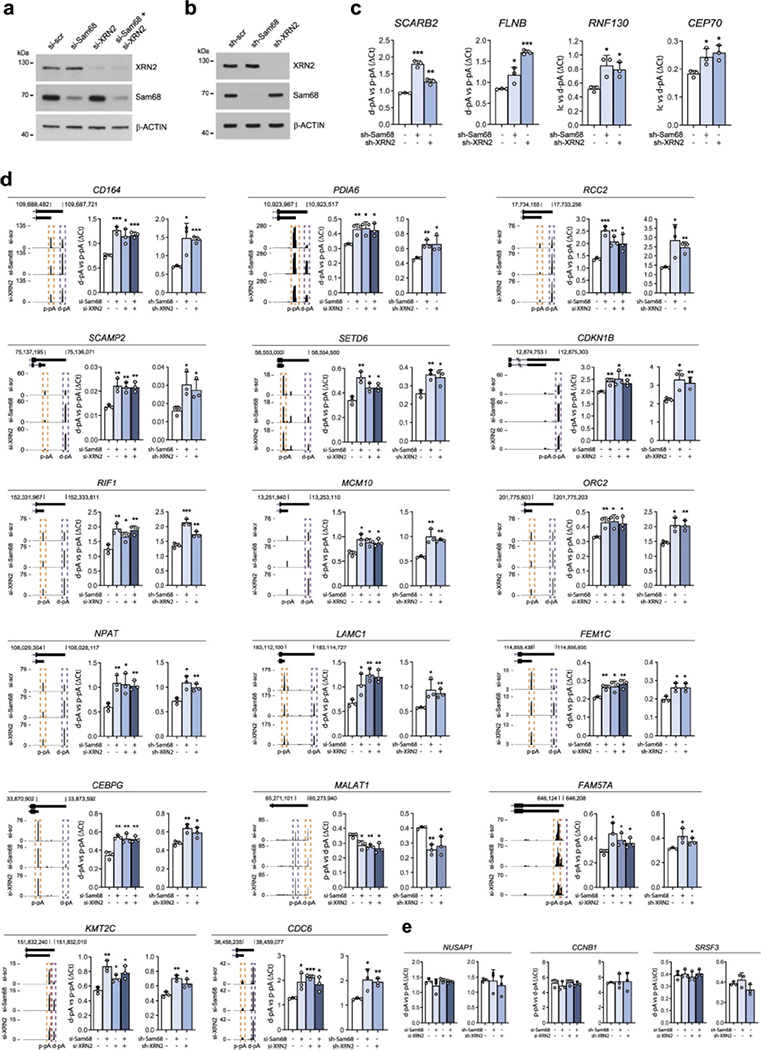
Extended Data Fig. 6: Genome-wide regulation of APA by XRN2 and Sam68 in PC cells (Related to Fig. 4).
From: The transcriptional terminator XRN2 and the RNA-binding protein Sam68 link alternative polyadenylation to cell cycle progression in prostate cancer
a,b, Representative Western blot analysis of LNCaP cells transiently (a) or stably (b) depleted for Sam68 and XRN2 (n = 3). β-actin was used as loading control, c-e, Bar graphs showing qRT-PCR analyses of pA usage evaluated in 24 representative genes undergoing APA regulation in LNCaP cells treated as in a and b. Fold change of d-pA relative to p-pA was calculated by the ΔCq method. In e, unvalidated genes are shown. Data represent mean ± SD of three biological replicates (c-e). In c-e, statistical significance was calculated by unpaired Student’s t-test, two-sided (exact p-values reported in source data). In the representation of panels, statistical value is reported as * P < 0.05; ** P < 0.01; *** P < 0.001. UCSC genome browser tracks showing APA regulation for each event analyzed is also shown on the right side of each graph. Purple and orange boxes indicate up- and down-regulated events, respectively.