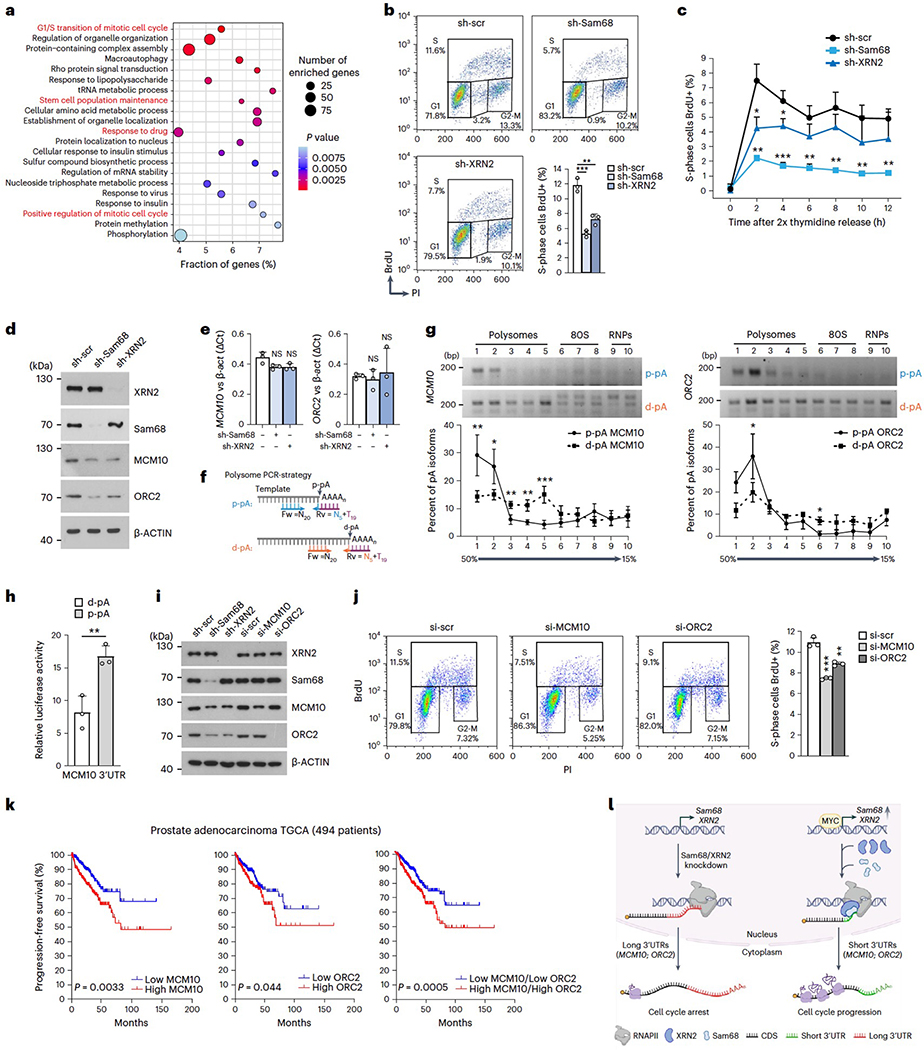
Fig. 7|. XRN2 and Sam68 promotes cell cycle progression through APA modulation.
a, Enrichment of Gene Ontology (GO) terms (dot plot) in genes regulated by 3’UTR-APA upon depletion of Sam68 or XRN2. Dot size and color indicate the number of genes and statistical significance (Fisher’s exact test, two-sided), respectively, b, Cytometric analyses showing DNA content versus BrdU incorporation upon stable depletion of Sam68 (sh-Sam68) and XRN2 (sh-XRN2) in LNCaP cells. The bar graph shows the percentage of BrdU-positive (S phase) cells, c. Percentage (mean + s.d.) of BrdU-positive LNCaP cells described in b at the indicated time points after release from G1/S synchronization. d,e, Western blot (d) and qPCR (e) analyses of MCM10 and ORC2 expression level in sh-Sam68 and sh-XRN2 LNCaP cells (n = 3). f, PCR strategy used to evaluate 3’UTR-APA isoforms distribution on a 15–50% sucrose gradient, g, sqPCR analysis of the indicated p-pA and d-pA isoform abundance within the polysomal and non-polysomal fractions obtained from sucrose gradient. The graphs show the densitometric analysis of the band signal in each fraction, expressed as a percentage of that detected in all fractions, h, Relative luciferase activity (Renilla/Firefly ratio) of long and short MCM10 3’UTR in LNCaP cells. i, Representative western-blot analysis (n = 3) of the indicated proteins performed in LNCaP cells depleted for the indicated genes, j, Cytometric analyses showing DNA content versus BrdU incorporation in control (si-scr), si-MCMlO and si-ORC2 LNCaP cells. The bar graph shows the percentage of S-phase BrdU-positive cells, k, Kaplan-Meier curves comparing progression-free survival of494 patients with PC (Prostate Adenocarcinoma, TCGA, PanCancer Atlas; https://www.cbioportal.org) stratified according to MCM10 (right), ORC2 (middle) and MCM10/ORC2 (left) expression level. I, Schematic model showing the impact of the functional interaction between Sam68 and XRN2 on cell cycle regulation. The Sam68/XRN2 complex promotes 3’UTR shortening of cell cycle-related genes, increasing their mRNA translation efficiency and cell proliferation. Conversely, Sam68/XRN2 knockdown induces 3’UTR lengthening, reduces translation efficiency of transcripts and causes cell cycle arrest. In b, e, h and j, the bar graphs represent the mean + s.d. In b, c, e, g, h and j, statistical significance was calculated by unpaired Student’sf-test, two-sided (n = 3; *P < 0.05, **P < 0.01,***P < 0.001; NS, not significant; exactPvalues are reported in the source data). In d and I, β-actin was used as loading control.