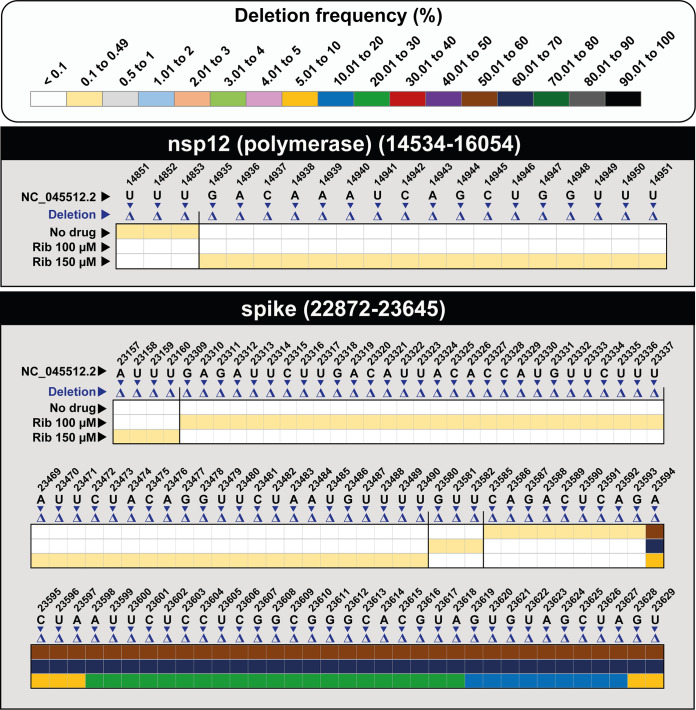
FIG 4.
Heatmap of SARS-CoV-2 deletions in SARS-CoV-2 populations grown in the absence (no drug) or presence of ribavirin (Rib), with a cutoff deletion frequency of 0.1%. Frequency values are color coded, as depicted in the top box. Data are presented in two blocks, one for the nsp12 (polymerase)-coding region (genomic residues 14,534 to 16,054) and another for the S-coding region (genomic residues 22,872 to 23,645). Only positions within a deletion are represented; deletions have been identified relative to NCBI reference sequence, GenBank accession no. NC_045512. The position, length, sequence context, frequency, and the number of affected haplotypes for each deletion are given in Fig. S2 at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX. Procedures are detailed in Materials and Methods.