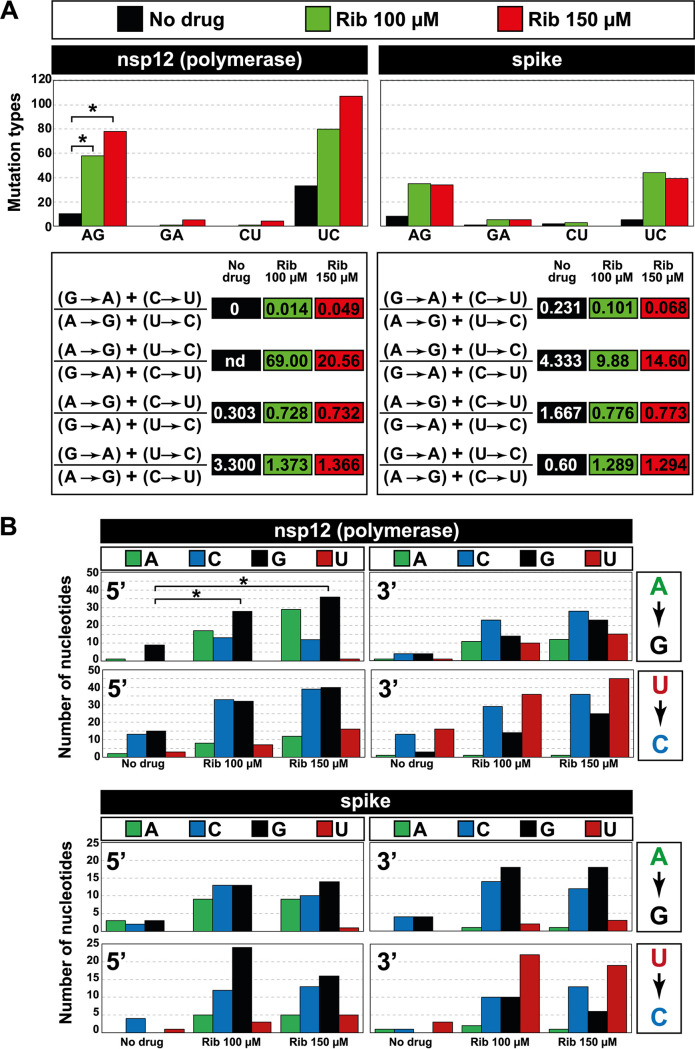
FIG 6.
Mutation types evoked by ribavirin on SARS-CoV-2 and neighbor preferences at the mutation types. (A) Mutation types evoked by ribavirin. The absence or presence of Rib is color coded in the top box. Left panels give data for the nsp12 (polymerase)-coding region, and right panels give data for the spike-coding region. Of the observed biases, the preference for A→G transitions reached statistical significance for the nsp12 (polymerase)-coding region (*, P < 0.05; proportion test). Mutation bias is reflected in transition type ratios, as depicted in the bottom panels; nd, not determined because the denominator for that ratio was zero. (B) Analysis of nucleotide preference at the 5′ side or 3′ side of the mutation sites. Coding region and nucleotide type are indicated in the top boxes, and mutation types are displayed in the boxes at the right of each panel. Absence or presence of ribavirin is given in the abscissae. The number of each type of nucleotide located at the 5′ side or 3′ side of the mutation site is written in ordinate; note the different scales for the two coding regions. The complete list of mutations on which the calculations are based is given in Table S3 in the supplemental material at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX.