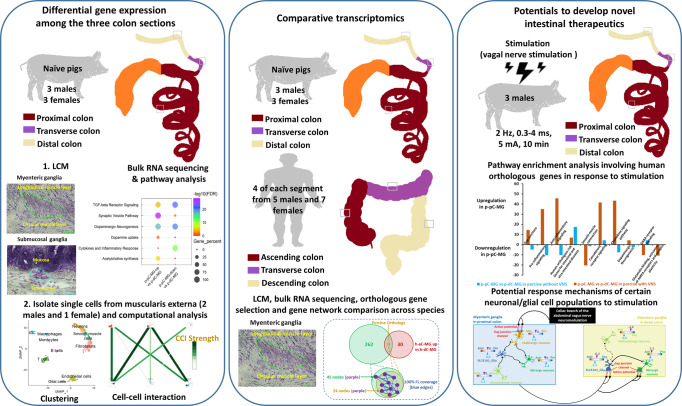
Fig. 1. Experimental design in the study.
The transcriptomes were profiled in myenteric and inner submucosal ganglia (MG, ISG) from proximal, transverse and distal colon (p-pC, p-tC, p-dC) of six naive pigs, and MG in parallel from ascending, transverse and descending colon (h-aC, h-tC, h-dC, 4 of each) from 12 patients (5 males and 7 females) using laser-capture microdissection (LCM) coupled with bulk RNA-seq analysis. Gene homology search was performed to obtain the orthologous genes between human and pig. The high-quality orthologous differentially expressed genes (DEG) derived from comparisons of MG between analogous colonic regions across species were extracted and the coverage of their functional linkages in the gene networks was used to reflect functional similarities across species. Based on the orthologous genes, the discrepancy of the region-specific gene programs was evaluated in MG and ISG after electrical stimulation of the celiac branch of the abdominal vagus nerve in the anesthetized pigs. Additionally, single-cell RNA-seq (scRNA-seq) was performed on the muscularis externa containing myenteric ganglia in three naive pigs (2 males and 1 female). Approximately, an average of 3200 cells (1400/3000/5300) from each p-pC individual, 3800 cells (2800/2200/6300) from each p-tC individual and 4100 cells (2400/5000/4920) from each p-dC individual were loaded on the 10× Genomics Chromium platform, generating 9 scRNA-seq datasets. This yielded an average of 1930 (735/2091/2965), 2318 (1993/1903/3057) and 2559 (1381/3918/2378) mapped single cells in each of p-pC, p-tC, and p-dC, respectively. The scRNA-seq was coupled with bulk gene expression deconvolution to identify cell-type marker genes and putative interactions among neurons and glia in MG of porcine colon and further revealed the regional transcriptomic heterogeneity in the enteric nervous system of the pigs with and without VNS.