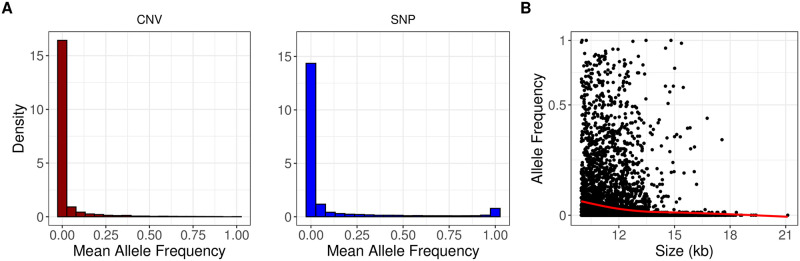
Fig 6. The impact of negative selection on deletions.
(A) The site frequency spectra of derived deletion alleles (on the left, n = 3,472) and derived SNP alleles (on the right, n = 57,307). Both plots represent frequency distributions of loci genotyped in all 50 ancient genomes (no missing data). Zero indicates lack of derived alleles. The two distributions are significantly different from each other (Kolmogorov-Smirnov test p < 10−15). (B) The size distribution (in kbps) of deletion events versus allele frequency. The red line shows the smoothing spline fit and indicates a negative correlation (Spearman correlation r = −0.33, p < 10−16). Both axes were log2-scaled.