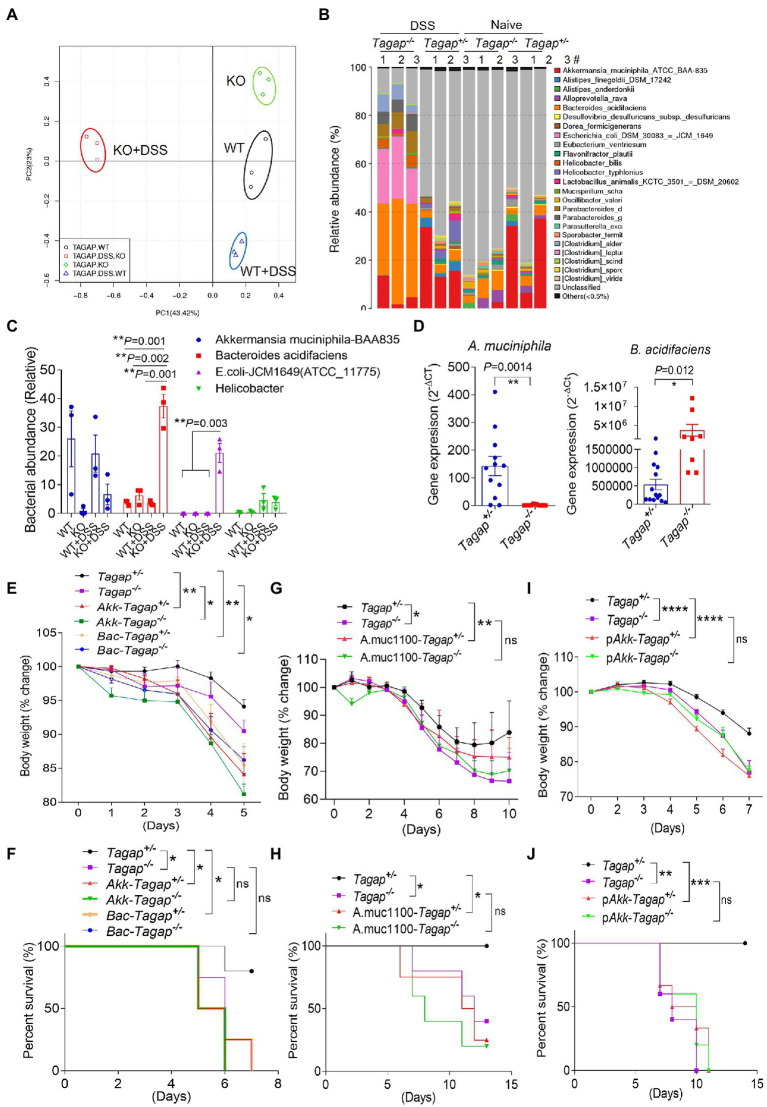
Figure 4.
TAGAP-deficient mice had altered microbiota in the gut. (A,B) Cluster analysis or diversity analysis of overall gut microbiota in the littermate control mice or TAGAP-deficient mice, n = 3. (C) Relative abundance of different bacterial strain in the feces of littermate control mice or TAGAP-deficient mice identified by 16S sequencing was shown, n = 3. (D) Feces from littermate control mice or TAGAP-deficient mice were collected and analyzed by real time PCR for the indicated bacterial strains, n = 12. (E,F) Littermate control mice or TAGAP-deficient mice were oral gavaged Akkermansia muciniphila or Bacteroides acidifaciens daily for 4 weeks (1 × 108/mouse/day), followed by 2.5% of DSS treatment for 5 days, and then changed to normal water. The weight loss (E) and survival data (F) were from separate experiments. (G,H) Littermate control mice or TAGAP-deficient mice were oral gavaged recombinant Amuc_1100 protein daily for 4 weeks (10 μg/mouse/day), followed by 2.5% of DSS treatment for 5 days. The weight loss (G) and survival data (H) were from separate experiments. (I,J) Littermate control mice or TAGAP-deficient mice were oral gavaged pasteurized A. muciniphila daily for 4 weeks (1 × 108/mouse/day), followed by 2.5% of DSS treatment for 5 days. The weight loss (I) and survival data (J) were from separate experiments, n = 5 for panels (E–J). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 based on 2-way ANOVA (E,G,I), Log-rank (Mantel-Cox) Test for panels (F,H,J) and unpaired T test (D). Data are representative of two independent experiments.