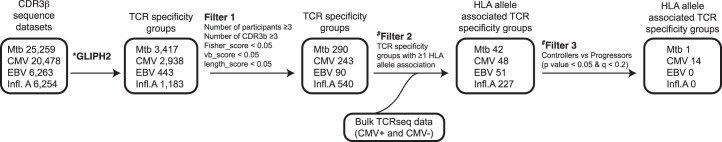
Extended Data Fig. 4. Differentially abundant CMV-specific TCR similarity groups in CMV+ persons compared to CMV- persons.
Analysis workflow used to measure the frequencies of GLIPH2 TCR similarity groupings from mycobacteria-reactive (Mtb) or CMV, EBV or Influenza-A (Infl.A)-specific CDR3β sequences in CMV+ and CMV- persons. GLIPH2 analysis was performed and the resulting GLIPH2 similarity groups were filtered initially using the criteria listed under Filter 1. We then selected TCR similarity groups with significant HLA allele associations in the CMV+/CMV- cohort (Filter 2). Finally, we identified similarity groups that were differentially abundant in CMV+ and CMV- participants bearing the associated HLA allele (Filter 3).