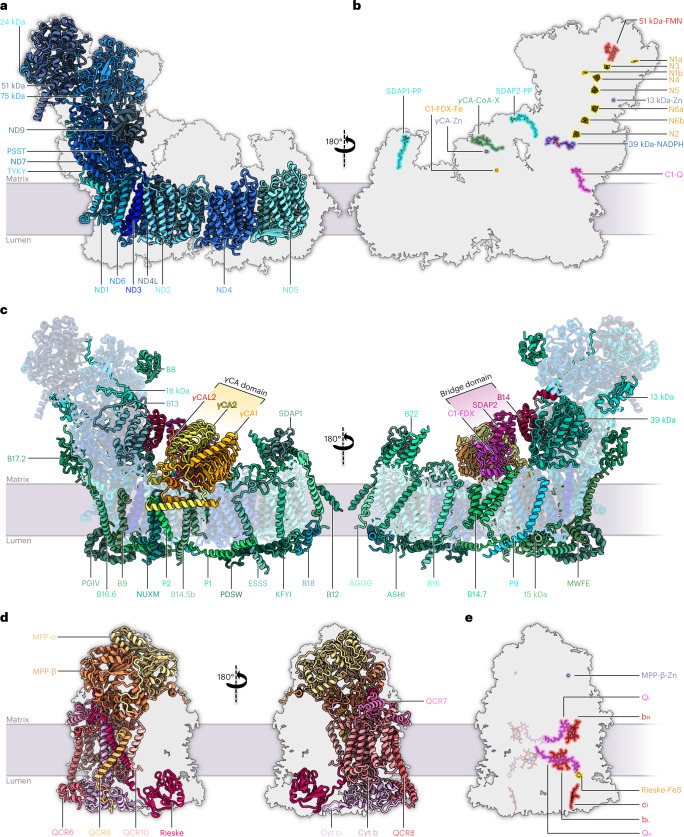
Fig. 2. Subunit composition of complex I and complex III2 within the Arabidopsis I + III2 supercomplex.
a–c, Atomic model of complex I, showing the 14 core subunits in shades of blue (a), cofactors bound to complex I (N, FeS clusters; Q, ubiquinone/ubiquinol; FMN, flavine mononucleotide) (b) and accessory subunits of mitochondrial complex I (c). Conserved accessory subunits are shown in shades of green, the subunits of the carbonic anhydrase (CA) domain in yellow and orange, and the subunits of the bridge domain in red and pink. The newly identified subunit P9 is light blue. Subunit nomenclature as for bovine complex I (ref. 80) except for non-conserved accessory subunits (for details, see Supplementary Table 5). d,e, Atomic model of complex III2, showing the structure of the ten subunits of one complex III monomer within the complex III dimer (for details, see Supplementary Table 6) from opposite directions (d) and bound cofactors of complex III2 (bH, bL, c1: haem groups attached to cytochrome b and c1; FeS: iron–sulphur cluster attached to the Rieske protein; Qi, Qo: quinone binding sites; Zn: zinc2+ bound to MPP-β) (e). For details and bound lipids, see Extended Data Fig. 2.