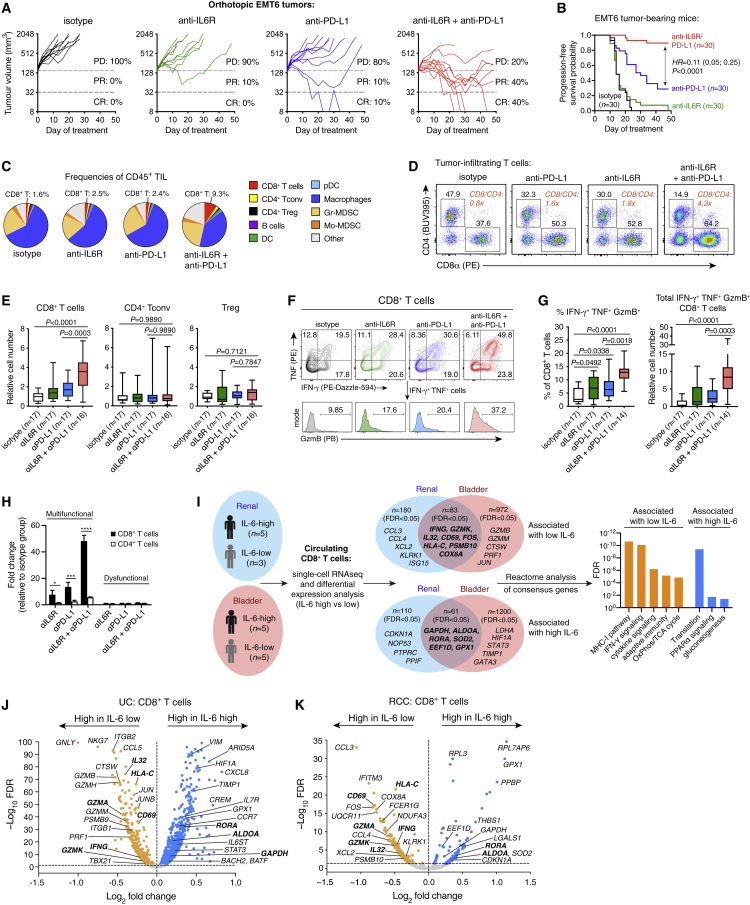
Figure 2.
IL-6 inhibits anti-PD-L1 efficacy and anti-tumor CTL response
(A–H) Treatment of EMT6 tumor-bearing mice with antibodies targeting PD-L1 and/or IL6R. (A) Individual tumor growth curves (n = 10 per group) from one of three independent studies. PD, progressive disease; PR, partial response; CR, complete response. (B) Progression-free survival (time to 5x increase in tumor volume) of mice pooled from three independent studies, analyzed using log rank test. (C) Cellular composition of CD45+ tumor-infiltrating leukocytes, from one of three experiments. (D) Balance of CD4+ and CD8+ T cells (CTLs) among total TCRβ+ tumor-infiltrating T cells. Data are concatenated from n = 5 mice per group, from one of three studies. (E) Relative abundance (normalized to tumor weight) of CTLs, conventional CD4+ T cells, and regulatory (CD4+ Foxp3+) T cells in EMT6 tumors, pooled from three experiments. Groups compared using one-way ANOVA with Holm-Sidak’s multiple comparisons test. (F) Effector phenotype of tumor-infiltrating CTLs after re-stimulation with PMA/ionomycin. (G) Frequencies of polyfunctional cells among tumor-infiltrating CTLs (left panel), and their absolute abundance relative to isotype control (right panel). Data pooled from three experiments. Groups compared using one-way ANOVA with Holm-Sidak’s multiple comparisons test. (H) Relative abundance (mean ± SEM) of polyfunctional CTLs and IFN-γ+TNF+ CD4+ T cells (multifunctional cells) vs. dysfunctional cells (IFN-γ− TNF− GzmB− CD8+ T cells, or IFN-γ− TNF− CD4+ T cells), from one of three experiments with n = 3–5 per group. Groups compared using one-way ANOVA with Holm-Sidak’s multiple comparisons test. ∗p = 0.0143, ∗∗∗p = 0.0001, ∗∗∗∗p < 0.0001.
(I–K) Single-cell RNA-seq analysis of pre-treatment peripheral blood CD8+ T cells from patients with RCC (from IMmotion150) or UC (from IMvigor210). Differential expression analysis was performed on CD8+ T cells from patients with high (>10 pg/mL) versus low plasma IL-6 (separate analyses for each cancer type). Genes identified as concordant in both cohorts (“consensus genes”) were evaluated for Reactome pathway enrichment (I). Volcano plots of differential gene expression are shown for UC and RCC samples in (J) and (K), respectively.