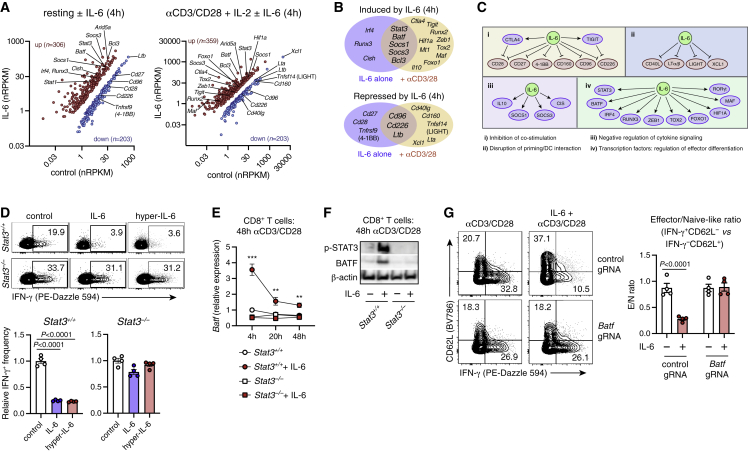
Figure 4.
IL-6 regulates CTLs via STAT3-dependent BATF induction
(A–C) RNA-seq analysis of WT naive CTLs stimulated with IL-6 +/− anti-CD3/CD28 antibodies for 4 h. (A) Differentially expressed genes (FDR <0.05 and absolute fold change >2).
(B) IL-6-regulated genes with potential roles in CTL differentiation, and their functional categorization (C).
(D) IFN-γ expression in WT or STAT3.ko CTLs (from CD4-Cre x Il6rloxP/loxP mice) activated with anti-CD3/CD28 +/− IL-6 or hyper-IL-6 for 3 days. Groups (mean ± SEM of n = 4 technical replicates) compared by t test; data representative of three independent experiments.
(E) Batf mRNA expression (qRT-PCR) in WT or STAT3.ko CTLs activated +/− IL-6. ∗∗p < 0.01, ∗∗∗p < 0.001 (WT vs. STAT3.ko; t tests). Data points indicate mean +/− SEM of n = 4 technical replicates, from one of two independent experiments.
(F) Western blot of BATF and p-STAT3 (Y705) in activated WT or STAT3.ko CTLs. Data represent one of two independent experiments.
(G) BATF CRISPR-ko or control CTLs were activated +/− IL-6 and analyzed on day 3 (groups compared by t test; mean ± SEM of n = 4 technical replicates). Data representative of two independent experiments.