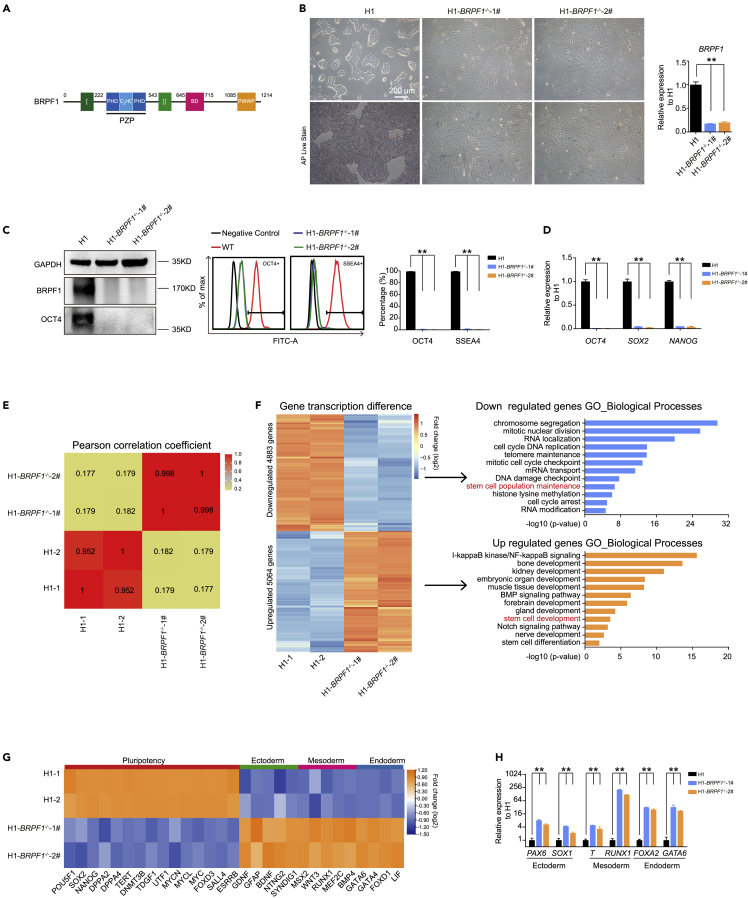
Figure 1.
BRPF1 is essential in human ESC pluripotency and self-renewal
(A) Overview of the BRPF1 domain architecture. I, enhancer of polycomb-I; II, enhancer of polycomb-II; PZP , PHD-zinc knuckle-PHD; BD, bromodomain; PWWP, Pro-Trp-Trp-Pro containing domain.
(B) Morphology of WT and BRPF1−/− H1 hESCs. Alkaline phosphatase (ALP) staining and expression of BRPF1 were examined in WT and two BRPF1−/− H1 hESC clones. The significance level was determined by unpaired two-tailed Student’s t-tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3). Scale bar, 200 μm.
(C). OCT4 and BRPF1 proteins were examined in WT and two BRPF1−/− H1 hESC clones by western blot or FACS analysis. The significance level was determined by unpaired two-tailed Student’s t tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3).
(D) Examination of these genes’ expression of OCT4, NANOG, and SOX2 in WT and two BRPF1−/− H1 hESC clones. The significance level was determined by unpaired two-tailed Student’s t tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3).
(E) Pearson’s correlation coefficient analysis of transcriptomes in WT and two BRPF1−/− H1 hESC clones.
(F) Heatmap and gene ontology (GO) analysis of up- or down-regulated genes in two BRPF1−/− H1 hESC clones compared with WT H1 hESCs.
(G) RNA-seq data of selected pluripotent or three germ layer genes in WT and two BRPF1−/− H1 hESC clones.
(H) Examination of the expression of lineage genes in WT and two BRPF1−/− H1 hESC clones. The significance level was determined by unpaired two-tailed Student’s t tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3). All error bars throughout the figure represent the SD from three independent replicates (n = 3).