Figure 3.
Decreased chromatin accessibility of stemness genes in BRPF1−/− H1 hESCs
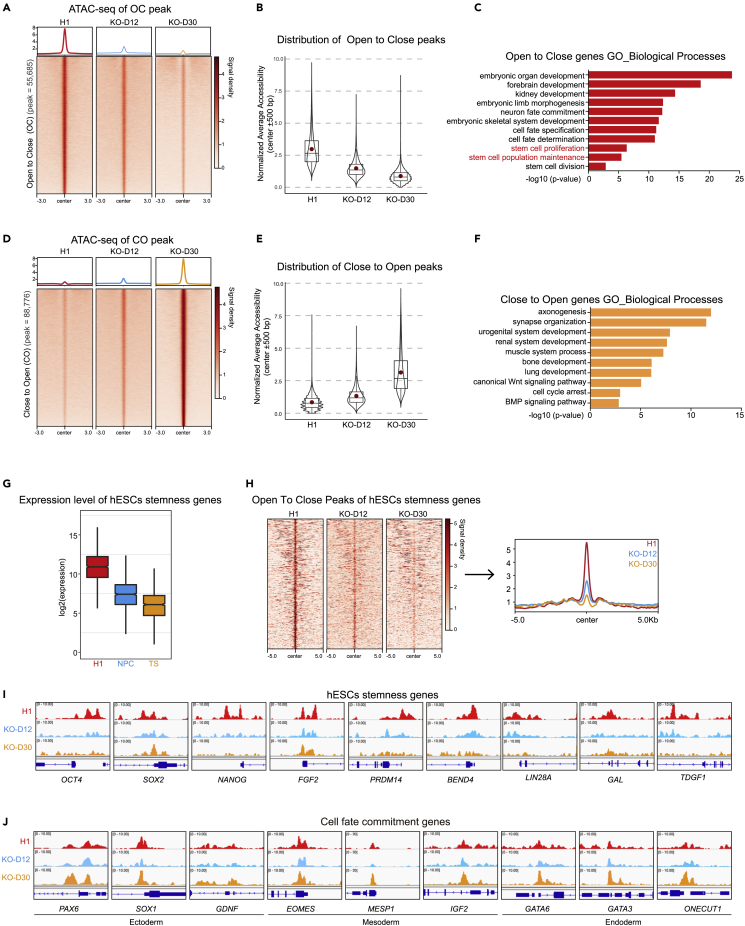
(A) Heatmap and signal densities of ATAC-seq data for OC accessible chromatin regions in WT and BRPF1−/− H1 hESCs (D12, D30). The signal around a ±3 kb window centered on OC peaks center in the indicated cells. OC, open to closed, indicates regions with reduced accessibility in BRPF1−/− H1 hESCs.
(B) Box-plot of signal densities of the ATAC-seq data for OC accessible chromatin regions in the indicated cells.
(C) GO analysis of genes associated with OC regions is shown.
(D) Heatmap and signal densities of ATAC-seq data for CO accessible chromatin regions in WT and BRPF1−/− H1 hESCs (D12, D30). The signal around a ±3 kb window centered on CO peaks center in the indicated cells. CO, close to open, indicates regions with increased accessibility in BRPF1−/− H1 hESCs.
(E) Box-plot of signal densities of the ATAC-seq data for CO accessible chromatin regions in the indicated cells.
(F) GO analysis of genes associated with CO regions is shown.
(G) Box-plot of transcription levels of selected stemness genes in H1 hESCs, H1 hESC-derived NPCs, and trophoblast stem (TS) cells.
(H) Heatmap and signal densities of ATAC data in regions associated with selected stemness genes. ATAC-seq signal around a ±5 kb window centered on OC peaks in the indicated cells.
(I and J) Genomic view of ATAC-seq data on selected genes with different functions in H1 and BRPF1−/− hESCs. These selected genes include (I) hESCs stemness genes and (J) cell fate commitment genes.