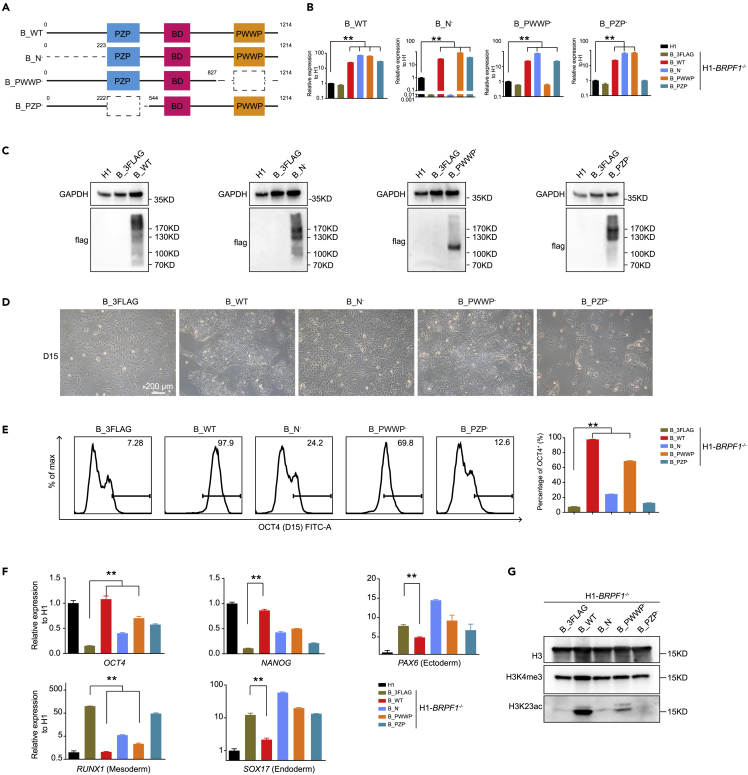
Figure 5.
Functional analysis of BRPF1 with the deletion of different histone binding modules
(A) Overview of wild-type BRPF1 and the BRPF1 mutants with domain architecture. Wild-type BRPF1 (B_WT); BRPF1 with N terminal deletion (B_N−); BRPF1 with PWWP module deletion (B_PWWP−); BRPF1 with PZP module deletion (B_PZP−). The dotted line indicates the deleted regions.
(B) qRT-PCR analysis of the expression with domain-specific primers. The significance level was determined by unpaired two-tailed Student’s t tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3).
(C) Western blot analysis of FLAG-tagged of overexpression of wild-type BRPF1 and each BRPF1 mutant in BRPF1−/− H1 hESCs. Overexpression of the negative control in BRPF1−/− H1 hESCs (B_3FLAG).
(D) Morphology of BRPF1−/− H1 hESCs overexpressing wild-type BRPF1 and each BRPF1 mutant. Scale bar, 200 μm.
(E) FACS analysis of OCT4 in BRPF1−/− H1 hESCs overexpressing wild-type BRPF1 and each BRPF1 mutant. The significance level was determined by unpaired two-tailed Student’s t tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3).
(F) qRT-PCR analysis of the expression of the pluripotent genes OCT4, NANOG, and cell fate commitment genes PAX6, RUNX1, and SOX17 in BRPF1−/− H1 hESCs overexpressing wild-type BRPF1 and each BRPF1 mutant. The significance level was determined by unpaired two-tailed Student’s t tests. ∗∗, p < 0.01. The data represent the mean ± SD (standard deviation) from three independent repeats (n = 3).
(G) Western blot analysis of H3K4me3 and H3K23ac in BRPF1−/− H1 hESCs overexpressing wild-type BRPF1 and each BRPF1 mutant. All error bars throughout the figure represent the SD (standard deviation) from three independent replicates (n = 3).