Abstract
Background:
Vitiligo is a common depigmentation disease characterized by progressive destruction and disappearance of epidermal melanocytes. Exosomes have been discovered to regulate the pigment status of melanocytes. We aimed to explore the role of exosomes from peripheral blood of vitiligo patients on melanogenesis.
Methods:
Human melanocytes cell line PIG1 was treated with exosomes from the healthy volunteers or exosomes from the vitiligo patients referred to the Department of Dermatology, Children’s Hospital Affiliated to Zhengzhou University, China and transfected with miR-21-5p mimic or inhibitor. Exosome labeling assay was used to assess whether exosomes were absorbed by melanocytes. Melanin content and tyrosinase activity assays were performed to investigate melanogenesis in melanocytes. The levels of melanogenesis-related genes and proteins were detected by RT-qPCR and western blot assays. Dual-luciferase reporter assay was performed to confirm the relationship between miR-21-5p and special AT-rich sequence binding protein-1 (SATB1).
Results:
Exosomes from peripheral blood of vitiligo patients were transferred into melanocytes and suppressed melanin content, tyrosinase activity and melanogenesis-related genes and proteins levels. Besides, miR-21-5p was highly expressed in exosomes from peripheral blood of vitiligo patients. The results of the gain- and loss-of-function experiments demonstrated that miR-21-5p inhibited the melanogenesis of melanocytes. Furthermore, miR-21-5p inhibitor abolished the inhibitory role of exosomes from peripheral blood of vitiligo patients. Subsequently, miR-21-5p directly targeted SATB1 in melanocytes. Furthermore, overexpression of SATB1 reversed the inhibitory roles of miR-21-5p mimic on melanin content, tyrosinase activity, and melanogenesis-related protein expression.
Conclusion:
Peripheral blood of vitiligo patients-derived exosomal miR-21-5p inhibited melanocytes melanogenesis via targeting SATB1.
Keywords: Vitiligo, Exosome, MIRN21 microRNA, Human, Melanogenesis, SATB1 protein
Introduction
Vitiligo is a common depigmentation disease characterized by progressive destruction and disappearance of epidermal melanocytes, with an estimated prevalence of 0.1–2% (1). It is extensively accepted that vitiligo seriously affects patients’ quality of life and causes serious psychological burden (2). The clinical treatment of vitiligo is limited because of its complex pathogenesis (1). In the genetic background, environmental factors, immunological factors and neuropsychiatric factors play a large part in the progression of vitiligo (3). Furthermore, melanocytes are the ultimate target of all pathogenic factors (4). Melanocytes are dendritic cells derived from the neural crest of the ectoderm. The main function of melanocytes is to produce melanin, which causes skin pigmentation. Therefore, improving the understanding of the pathogenesis of vitiligo is essential for developing more effective treatment strategies.
In recent years, exosomes have attracted numerous research interests worldwide. Exosomes are lipid bilayer membrane vesicles 30–150 nm in diameter (5). Exosomes are important mediators of intercellular communication and participate in biological processes such as immune response, apoptosis, angiogenesis and inflammation (6). The changes of exosome contents reflect the physiological and pathological conditions of donor cells. At the same time, exosomes also transport their genetic materials, such as lipids, proteins, mRNA, microRNAs (miRNAs), and DNA, into target cells, regulate gene expression and affect the activity of related pathways in target cells (7). Exosomes regulate the pigment status of melanocytes by changing gene expression and enzyme activity (8, 9). Exosomes affect the balance of CD8+, Treg, and Th17 cells, which contribute to the destruction of autoimmune tolerance in vitiligo (10). In addition, exosomes act as communication mediators between keratinocytes and melanocytes in the melanogenesis pathway and participate in the transport of melanosomes (11). Exosomes also regulate melanocyte survival during melanin production and protein expression of enzymes such as tyrosinase, tyrosinase associated protein 1 (TRP1), tyrosinase associated protein 2 (TRP2), and small eye associated transcription factor (MITF) (12). As is known, exosomes are widely found in body fluids, including blood, urine and saliva (13). However, it remains unknown whether exosomes from peripheral blood of vitiligo patients are associated with melanogenesis.
MiRNAs are a kind of endogenous small non-coding RNA with 19–22 nucleotides in length, which down-regulate gene expression after transcription by targeting binding to the 3’-untranslated regions (3’-UTR) (14). MiRNAs were abnormally expressed in vitiligo patients (15, 16). Besides, miRNAs play important roles in the development of melanogenesis (17).
We aimed to explore the role of exosomes from peripheral blood of vitiligo patients in melanogenesis.
Material and Methods
Human samples
Human peripheral blood samples (5 mL) from 10 healthy volunteers and 10 patients who had been diagnosed with vitiligo were collected from Children’s Hospital Affiliated to Zhengzhou University. All participants signed the informed consent before inclusion in the study. The project protocol was approved by the Ethics Committee of Children’s Hospital Affiliated to Zhengzhou University.
Isolation of exosomes
Exosomes were isolated from peripheral blood using the ExoQuick Kit (SBI, USA). In brief, the collected blood was centrifuged at 4 °C at 3000 g for 15 min. Then, the supernatant was filtered through a 0.22 μm filter, and incubated with precipitation solution at 4 °C for 30 min. The mixture was subsequently centrifuged at 1500 g for 30 min. The cream precipitate (exosomes particle) was resuspended in 200 μL phosphate-buffered saline (PBS, Gibco, USA). The exosome particle suspension was stored at −80 °C.
Characterization of exosomes
Transmission electron microscopy (TEM) was used to identify morphology. In brief, 20 μL suspension resuspended by PBS was dropped onto a diameter copper network, and placed at room temperature for 2 min. Then, 30 μL phosphotungstic acid solution was added to samples for 5 min. The morphological characteristics of the isolated exosomes were viewed by JEM-1-11 microscope (Philips, Netherlands).
Nanoparticle tracking analysis (NTA) was used to analyze the particle diameter distribution of the isolated exosomes using the NanoSight LM10 instrument (Malvern Instruments, Ltd., UK).
Cell culture
Human melanocyte cell line PIG1 was purchased from the American Type Culture Collection (ATCC, USA). PIG1 cells were cultured in DMEM medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA), 100 μg/ml penicillin (Sigma-Aldrich, USA), and 100 μg/mL streptomycin (Sigma-Aldrich, USA) at 37 °C with 5% CO2.
Cell treatment and transfection
To explore the roles of exosomes on melanogenesis in PIG1 cells, PIG1 cells were treated with 100 μg/mL exosomes derived from the healthy volunteers (Normal-exo) or 100 μg/mL exosomes derived from the vitiligo patients (Patients-exo) for 24 h.
The miR-21-5p mimic, miR-21-5p inhibitor and corresponding negative control were synthesized from GenePharma (Shanghai, China). The sequences for cell transfection were listed in Table 1. For overexpression of SATB1 (SATB1 OE), the cDNA of SATB1 was cloned into pcDNA3.1 vector. pcDNA3.1 vector was considered as the overexpression of negative control (NC OE). When cells reached 70% confluence, PIG1 cells were transfected with oligonucleotide or plasmid using the Lipofectamine RNAiMAX reagent or Lipofectamine 2000 (Invitrogen, USA).
Table 1:
The sequences for cell transfection
| Gene | Sequences (5’-3’) |
|---|---|
| miR-21-5p mimic | UAGCUUAUCAGACUGAUGUUGA |
| miR-21-5pinhibitor | UCAACAUCAGUCUGAUAAGCUA |
| NC mimic | UCACAACCUCCUAGAAAGAGUAGA |
| NC inhibitor | UCACAACCUCCUAGAAAGAGUAGA |
Exosome labeling assay
Normal-exo and patients-exo were labeled with PKH67 green CM-Dil dye (Sigma-Aldrich, USA) for 5 min. Then, PIG1 cells were incubated with PKH67-labeled Normal-exo or Patients-exo for 24 h. After incubation, PIG1 cells were fixed with 4% paraformaldehyde (Beyotime, China) and cell nuclei was stained with DAPI (Beyotime, China). The images were observed by a fluorescence microscope (Nikon, Japan).
Melanin content assay
After treatment, PIG1 cells were washed with PBS twice and dissolved in 1 mol/L NaOH (Sigma-Aldrich, USA) supplemented with 10% DMSO (Sigma-Aldrich, USA) at 80°C for 30 min. The absorbance of melanin was detected by a microplate reader (Bio-Rad, China) at 475 nm.
Tyrosinase activity assay
After treatment, PIG1 cells were washed with PBS twice and suspended in 1% TritonX-100 (Sigma-Aldrich, USA). Then, PIG1 cells were disrupted by sonication, and the cell lysate was centrifuged at 13 000 g for 10 min. The supernatant was incubated with 2 mg/mL levodopa at 37 °C for 30 min. The absorbance was detected by a microplate reader (Bio-Rad, China) at 475 nm.
Dual-luciferase reporter assay
Online database TargetScan was used to predict the putative binding sites between miR-21-5p and SATB1. The wild type of SATB1 was cloned into pGL3-Firefly-Renilla vector (Promega, USA). The binding site of 3’-UTR of SATB1 was mutated from CAAGCAAUUCAUACUAUAAGCUU to CAAGCAAUUCAUACUUAUUCGAU for miR-21-5p. Then, the mutant type of SATB1 was also cloned into pGL3-Firefly-Renilla vector (Promega, USA). PIG1 cells were plated into 24-well plates, and cotransfected with luciferase plasmids and mimic or inhibitor for 24 h. Following transfection, the luciferase activity was measured by the Dual-Luciferase Reporter Assay (Promega, USA) according to the manufacturer’s instruction.
Western blot
Total protein was isolated from exosomes or PIG1 cells using RIPA plus PMSF (Beyotime, China). The protein concentration was detected by the BCA Kit (Beyotime, China). The protein samples were separated by SDS-PAGE gel electrophoresis, and then transferred to PVDF membranes. After blocking, the membranes were incubated with the specific primary antibodies anti-CD63 (cat: ab193349, 1:1000, Abcam, USA); anti-TSG101 (cat: #72312, 1:1000, CST, USA); anti-MITF (cat: #12590, 1:1000, CST, USA); anti-Tyrosinase (cat: ab170905, 1:1000, Abcam, USA); anti-TRP1 (cat: ab235447, 1:1000, Abcam, USA); anti-TRP2 (cat: ab234901, 1:1000, Abcam, USA); anti-SATB1 (cat: ab109122, 1:1000, Abcam, USA); anti-GAPDH (cat: #5174, 1:2000, CST, USA) at 4°C overnight. The horseradish peroxidase-labeled secondary antibody (1:1000, Abcam, USA) was incubated for 2 h at room temperature. The ECL kit (Beyotime, China) was used to visualize the membrane. The proteins were analyzed with Image J software.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from exosomes or PIG1 cells using TRIzol (Qiagen, USA). The RNA concentration was detected by NanoDrop (Thermo, USA). Then, total RNA was reverse-transcribed to cDNA by PrimeScript RT kit (Takara, Japan). RT-qPCR was implemented on ABI Real-Time PCR System (Applied Biosystems, USA) using SYBR Green Master Mix or TaqMan MicroRNA Assay Kit (Takara, Japan). MiRNA or mRNA expression levels were calculated using 2−ΔΔCT method and were normalized to U6 or β-actin. The primer sequences used in the study are listed in Table 2.
Table 2:
Primer sequences of genes in RT-qPCR assay
| Gene | Forward Primer (5′-3′) | Reversed Primer (5′-3′) |
|---|---|---|
| MITF | CGAGCGTCCTGTATGCAGAT | TCAAGTTTCCCGAGACAGGC |
| Tyrosinase | CTTCACAGGGGTGGATGACC | TGGTCCCCAAAAGCCAAACT |
| TRP1 | TGGTATGTGTTGCCCAGACC | ATGGGGATACTGAGGGCTGT |
| TRP2 | TTGCTTGGGCTGCAAAATCC | CTTGCTGAGAGCCACAGACA |
| PPP1R3B, | CTCACCAAAGCCCAGCAAAC | TAGCGGGTCATCGAATTCCG |
| RALGPS2 | TCCCATGTTGCCTCATGTCC | TCTCTGGAAGCAGCAGAACG |
| PAN3 | TCGTTGTCTGCTGGGTCTTC | GGCACTAGAAGGCACCATGT |
| SOX5 | GTCAGCTGCCGCCATTAATG | ATAGCTGAAGCCTGGAGGGA |
| XKR6 | TACAGAGGGGCCATCATCCA | AGAAGTCTGTTCCGCCATGG |
| KLF6 | CCACTTGAAAGCACACCAGC | CCTGTCACAGTGGGAGCATT |
| STRN | GTTTTACGAGGCCCTCTGCT | AGATCCACAGAGGCAGGGAT |
| GID4 | AGGGGAACTCGTACGACGTA | TGCATCCCACTTGCGAGTTA |
| SATB1 | GCTGCCAGTTTTCTGTGTGG | TGCCATTTCGATCAGCTGGT |
| NFIA | GCACATCCTCTACGAGCTCC | TCAGTGTGGTTGGAGATGGC |
| miR-21-5p | GCGCGTAGCTTATCAGACTGA | AGTGCAGGGTCCGAGGTATT |
| GAPDH | GAGAAGGCTGGGGCTCATTT | AGTGATGGCATGGACTGTGG |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
The data were presented as means ± SEM. The differences among multiple groups were analyzed using a one-way ANOVA followed by a Tukey’s post hoc test and the differences between two groups were analyzed using a paired student’s t-test. Differences were considered statistically significant at the level of P < 0.05.
Results
Exosomes from peripheral blood of vitiligo patients inhibit the melanogenesis of melanocytes
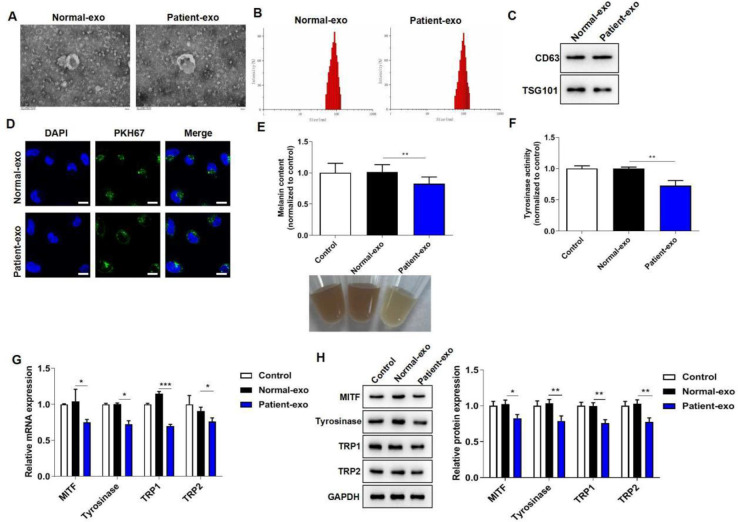
Exosomes were isolated from peripheral blood of vitiligo patients and healthy volunteers, and the characterization of the isolated particles was identified by TEM and NTA. The data showed that both Normal-exo and Patient-exo were spherically shaped vesicles of 30–150 nm in diameter (Fig. 1A–B). Moreover, western blot assay suggested that both Normal-exo and Patient-exo were positive for CD63 and TSG101 expression (Fig. 1C). These results indicated that Normal-exo and Patient-exo were consistent with the characteristics of exosomes. Then, melanocytes were incubated with PKH67-labeled Normal-exo or Patients-exo for 24 h. A fluorescence microscope observed that both Normal-exo and Patient-exo were taken up by melanocytes (Fig. 1D).
Fig. 1:
Exosomes from peripheral blood of vitiligo patients inhibit the melanogenesis of melanocytes. (A) The morphological characteristics of Normal-exo and Patient-exo were observed by TEM. (B) The diameter distribution of Normal-exo and Patient-exo was detected by NTA. (C) Western blot assay was used to detect CD63 and TSG101 expression. (D) The uptake ability of exosomes by melanocytes was observed by fluorescence microscope. (E) Melanin content in melanocytes were detected by a microplate reader. (F) Tyrosinase activity in melanocytes were detected by a microplate reader. (G) The mRNA levels of MITF, tyrosinase, TRP1 and TRP2 were assessed by RT-qPCR assay. (H) The protein levels of MITF, tyrosinase, TRP1 and TRP2 were assessed by western blot assay.
*P < 0.05, **P < 0.01, ***P < 0.001
To explore the role of exosomes from peripheral blood of vitiligo patients on melanogenesis, PIG1 cells were co-cultured with PBS, Normal-exo or Patient-exo for 24 h. Compared to Normal-exo, Patient-exo treatment significantly suppressed melanin content and tyrosinase activity in melanocytes (Fig. 1E–F). Moreover, the results of RT-qPCR and western blot assays showed that Patient-exo dramatically down-regulated the mRNA and protein levels of MITF, tyrosinase, TRP1 and TRP2 (Fig. 1G–H). The above results indicated that melanocytes treated with exosomes from peripheral blood of vitiligo patients inhibited melanogenesis.
miR-21-5p is highly expressed in exosomes from peripheral blood of vitiligo patients and inhibits the melanogenesis of melanocytes
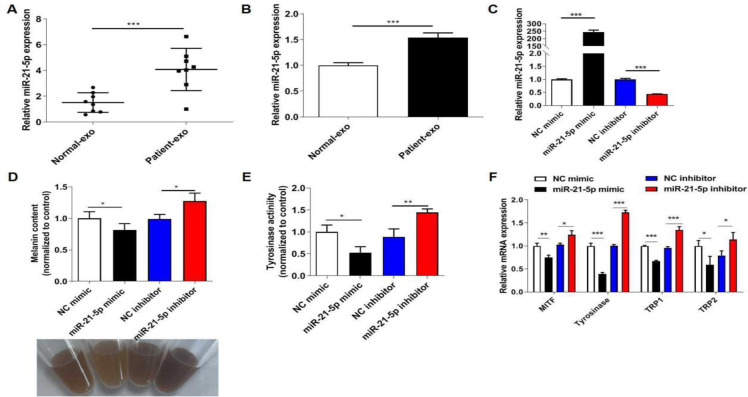
It’s reported that miR-21-5p level is increased in vitiligo patients and participates in the progression of vitiligo (15, 18). RT-qPCR assay suggested that Patient-exo had a higher expression of miR-21-5p than Normal-exo (Fig. 2A). Additionally, melanocytes treated with Patient-exo also increased miR-21-5p expression (Fig. 2B). To clarify the role of miR-21-5p on melanogenesis, miR-21-5p was knocked down or overexpressed in PIG1 cells. The transfection efficiency of miR-21-5p mimic and inhibitor in PIG1 cells was evidenced by RT-qPCR assay (Fig. 2C). Then, miR-21-5p mimic notably suppressed melanin content and tyrosinase activity in melanocytes, while miR-21-5p inhibitor promoted melanin content and tyrosinase activity (Fig. 2D–E). Furthermore, miR-21-5p mimic down-regulated the mRNA levels of MITF, tyrosinase, TRP1 and TRP2, while miR-21-5p inhibitor had the opposite roles (Fig. 2F). All these data suggested that the inhibitory role of Patient-exo on melanogenesis was partly ascribed to the highly expressed miR-21-5p.
Fig. 2:
miR-21-5p is highly expressed in exosomes from peripheral blood of vitiligo patients and inhibits the melanogenesis of melanocytes. (A) RT-qPCR assay was used to detect miR-21-5p expression between Patient-exo and Normal-exo. (B) RT-qPCR assay was used to detect miR-21-5p expression in PIG1 cells with Patient-exo or Normal-exo treatment. (C) The transfection efficiency of miR-21-5p mimic and inhibitor in PIG1 cells was evidenced by RT-qPCR assay. (D) Melanin content in melanocytes was detected by a microplate reader. (E) Tyrosinase activity in melanocytes was detected by a microplate reader. (F) The mRNA levels of MITF, tyrosinase, TRP1 and TRP2 were assessed by RT-qPCR assay. *P < 0.05, **P < 0.01, ***P < 0.001
Exosomal miR-21-5p from peripheral blood of vitiligo patients inhibits the melanogenesis of melanocytes
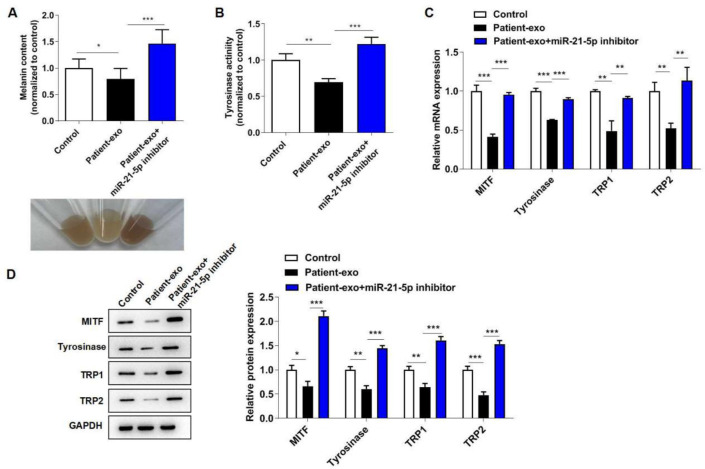
To explore whether miR-21-5p participated in exosomes-mediated melanocytes melanogenesis, PIG1 cells were co-cultured with Patient-exo and transfected with miR-21-5p inhibitor for 24 h. As expected, Patient-exo treatment significantly suppressed melanin content and tyrosinase activity in melanocytes, while further miR-21-5p inhibitor transfection increased melanin content and tyrosinase activity (Fig. 3A–B). Moreover, miR-21-5p inhibitor abolished the inhibitory role of Patient-exo on MITF, tyrosinase, TRP1 and TRP2 levels (Fig. 3C–D). These findings unraveled that exosomal miR-21-5p was associated with melanogenesis.
Fig. 3:
Exosomal miR-21-5p from peripheral blood of vitiligo patients inhibits the melanogenesis of melanocytes. PIG1 cells were co-cultured with Patient-exo and transfected with miR-21-5p inhibitor for 24 h. (A) Melanin content in melanocytes was detected by a microplate reader. (B) Tyrosinase activity in melanocytes was detected by a microplate reader. (C) The mRNA levels of MITF, tyrosinase, TRP1 and TRP2 were assessed by RT-qPCR assay. (D) The protein levels of MITF, tyrosinase, TRP1 and TRP2 were assessed by western blot assay. *P < 0.05, **P < 0.01, ***P < 0.001
miR-21-5p directly targets SATB1 in melanocytes
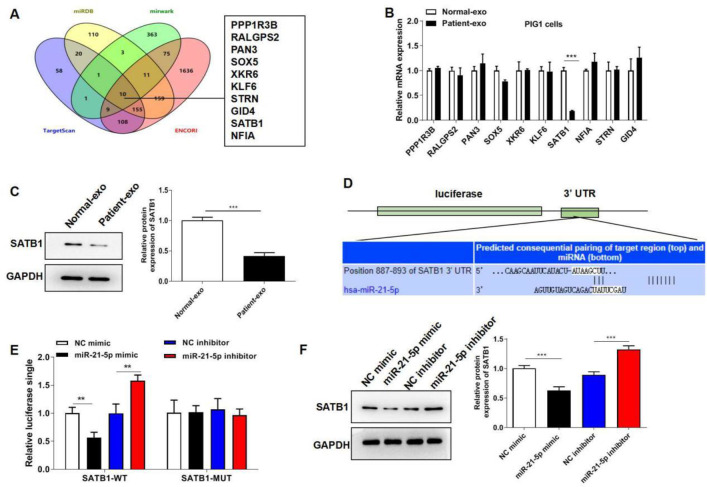
To further illuminate the regulatory mechanism of exosomal miR-21-5p in the progression of melanocytes melanogenesis, online databases (miRDB, miRWalk, TargetScan, and ENCORI StarBase) were used to predict the target genes of miR-21-5p. Venn diagram showed that PPP1R3B, RALGPS2, PAN3, SOX5, XKR6, KLF6, STRN, GID4, SATB1 and NFIA were the 10 common target genes in the four databases (Fig. 4A). Besides, RT-qPCR assay suggested that SATB1 mRNA level was significantly down-regulated in PIG1 cells cultured with Patient-exo (Fig. 4B). Western blot also showed that Patient-exo decreased SATB1 protein level (Fig. 4C). The potential binding sites of miR-21-5p with 3’-UTR of SATB1 were shown in Fig. 4D. miR-21-5p mimic significantly decreased the luciferase activity of the wild type SATB1 rather than the mutant type of SATB1 (Fig. 4E). Western blot further showed that miR-21-5p mimic dramatically suppressed SATB1 level, while miR-21-5p inhibitor significantly expedited SATB1 level (Fig. 4F). These data indicated that miR-21-5p negatively modulated SATB1 expression by directly binding to its 3’-UTR.
Fig. 4:
miR-21-5p directly targets SATB1 in melanocytes. (A) Online databases were used to predict the target genes of miR-21-5p. (B) RT-qPCR assay was performed to investigate SATB1 mRNA level in PIG1 cells with Patient-exo and Normal-exo treatment. (C) Western blot assay was performed to investigate SATB1 protein level in PIG1 cells with Patient-exo or Normal-exo treatment. (D) The potential binding sites of miR-21-5p with 3’-UTR of SATB1. (E) Dual-luciferase reporter assay was performed to confirm the relationship between miR-21-5p and SATB1. (F) Western blot assay was performed to investigate SATB1 protein level in PIG1 cells with miR-21-5p mimic or inhibitor transfection. *P < 0.05, **P < 0.01, ***P < 0.001
miR-21-5p inhibits the melanogenesis of melanocytes via targeting SATB1
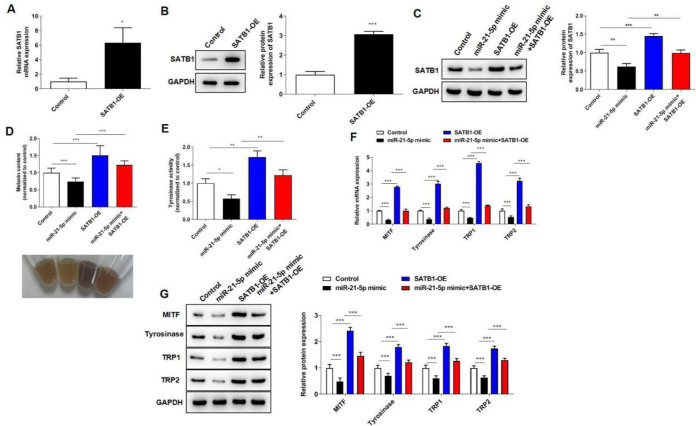
To further investigate the regulatory effect of the miR-21-5p/SATB1 axis in melanocytes melanogenesis progression, PIG1 cells were transfected with miR-21-5p mimic and SATB1 overexpression plasmid for 24 h. The transfection efficiency of SATB1 overexpression plasmid in PIG1 cells was shown in Fig. 5A–B. Besides, miR-21-5p mimic suppressed SATB1 protein expression, and overexpression of SATB1 further increased SATB1 protein expression (Fig. 5C). Functionally, overexpression of SATB1 reversed the inhibitory roles of miR-21-5p mimic on melanin content, tyrosinase activity, and melanogenesis-related protein expression (Fig. 5D–G). The data highlighted the importance of the miR-21-5p/SATB1 signaling axis in melanogenesis.
Fig. 5:
miR-21-5p inhibits the melanogenesis of melanocytes via targeting SATB1. PIG1 cells were transfected with miR-21-5p mimic and SATB1 overexpression plasmid for 24 h. (A) The transfection efficiency of SATB1 overexpression plasmid in PIG1 cells was confirmed by RT-qPCR assay. (B) Western blot assay showed the transfection efficiency of SATB1 overexpression plasmid in PIG1 cells. (C) Western blot assay showed SATB1 protein expression in PIG1 cells. (D) Melanin content in melanocytes was detected by a microplate reader. (E) Tyrosinase activity in melanocytes was detected by a microplate reader. (F) The mRNA levels of MITF, tyrosinase, TRP1 and TRP2 were assessed by RT-qPCR assay. (G) The protein levels of MITF, tyrosinase, TRP1 and TRP2 were assessed by western blot assay. *P < 0.05, **P < 0.01, ***P < 0.001
Discussion
It is accepted that pathological factors such as autoimmune factors, oxidative stress factors, neuropsychiatric factors and genetic predisposition factors cause melanocyte dysfunction (19). Melanin is synthesized in the melanosomes of melanocytes. The phenotypic diversity of skin pigmentation is closely related to the diversity of the number, size, composition and distribution of melanosomes (20). Defects in melanin production affect the physiology of multiple organ systems and cause disease. Vitiligo is one of the most common depigmentation skin diseases characterized by abnormal regulation of melanocytes melanogenesis (21). For example, the accumulation of abnormal melanosome structures in vitiligo melanocytes is associated with polymorphism in the tyrosinase gene (22). Furthermore, evidence has shown that a positive CD8+T cell could be isolated from peripheral blood of patients with vitiligo, which kills melanocytes in vitro (23). In order to observe more intuitively the influence of exosomes from peripheral blood of vitiligo patients on melanocytes, we mainly studied the internalization process of exosomes, tyrosinase activity, melanin content and other indicators.
Melanogenesis is associated with exosomes (24–26). For instance, exosomes from keratinocytes suppressed melanin production in melanoma cells (24). Besides, exosomes from amniotic stem cells inhibited melanogenesis and induced melanosome degradation (25). Exosomes from milk inhibited melanogenesis progression (8). To further explore the role of the uptake of exosomes from peripheral blood of vitiligo patients by melanocytes on melanogenesis, the exosomes were co-cultured with PIG1 for 24 h. The data showed that exosomes from peripheral blood of vitiligo patients inhibited melanocytes melanogenesis.
MiRNAs provide important information for related diseases, and become potential biomarkers in the development of vitiligo. Shang Z et al. used miRNA array to analyze the differentially expressed miRNAs in peripheral blood from vitiligo patients, and found four abnormally expressed miRNAs (miR-1238-3p, miR-202-3p, miR-630 and miR-766-3p) (16). Zhang Z et al. also screened for miRNAs in the peripheral blood mononuclear cells of non-segmental vitiligo patients, and showed miR-20a-5p was the molecular marker for stage assessment of nonsegmental vitiligo (26). These studies further highlighted the importance of miRNAs in patients with vitiligo. Besides, serum level of miR-21-5p was reported to be increased in vitiligo patients and associated with vitiligo area and severity index score (15). MiR-21-5p suppressed melanocytes destruction and regulated the balance of Treg/Teff to alleviate vitiligo (18). Our results revealed that miR-21-5p was highly expressed in exosomes from peripheral blood of vitiligo patients and inhibited the melanogenesis of melanocytes.
The present study also showed that miR-21-5p negatively modulated SATB1 expression by directly binding to its 3’-UTR. SATB1 was considered as a predictor of poor prognosis in cutaneous malignant melanoma (27). Furthermore, overexpression of SATB1 promoted the progression of melanoma (27). In the present study, we found that overexpression of SATB1 reversed the inhibitory roles of miR-21-5p mimic on melanin content, tyrosinase activity, and melanogenesis-related proteins expression. To the best of our knowledge, the present study was the first to identify the role of SATB1 on melanocytes.
Conclusion
The present study clarified the important signaling axis involved in the associated mechanism between vitiligo and melanocytes. Exosomal from peripheral blood of vitiligo patients suppressed melanocytes melanogenesis via targeting SATB1.
Journalism Ethics considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgements
No funding was received in this study.
Footnotes
Conflict of interest
The authors declare that there is no conflict of interest.
References
- 1.Frisoli ML, Essien K, Harris JE. (2020). Vitiligo: Mechanisms of Pathogenesis and Treatment. Annu Rev Immunol, 38:621–648. [DOI] [PubMed] [Google Scholar]
- 2.Thakur V, Bishnoi A, Vinay K, Kumaran SM, Parsad D. (2021). Vitiligo: Translational research and effective therapeutic strategies. Pigment Cell Melanoma Res, 34:814–826. [DOI] [PubMed] [Google Scholar]
- 3.Al Abadie MS, Gawkrodger DJ. (2021). Integrating neuronal involvement into the immune and genetic paradigm of vitiligo. Clin Exp Dermatol, 46:646–650. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Li S, Li C. (2021). Mechanisms of melanocyte death in vitiligo. Med Res Rev, 41:1138–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalluri R, LeBleu VS. (2020). The biology, function, and biomedical applications of exosomes. Science, 7;367(6478):eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tavasolian F, Hosseini AZ, Rashidi M, et al. (2021). The Impact of Immune Cell-derived Exosomes on Immune Response Initiation and Immune System Function. Curr Pharm Des, 27:197–205. [DOI] [PubMed] [Google Scholar]
- 7.Barile L, Vassalli G. (2017). Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol Ther, 174:63–78. [DOI] [PubMed] [Google Scholar]
- 8.Bae IS, Kim SH. (2021). Milk Exosome-Derived MicroRNA-2478 Suppresses Melanogenesis through the Akt-GSK3β Pathway. Cells, 10(11):2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melnik BC, John SM. (2020). MicroRNA-21-Enriched Exosomes as Epigenetic Regulators in Melanomagenesis and Melanoma Progression: The Impact of Western Lifestyle Factors. Cancers (Basel), 29;12(8):2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong PM, Yang L, Yang L, et al. (2020). New insight into the role of exosomes in vitiligo. Autoimmun Rev, 19:102664. [DOI] [PubMed] [Google Scholar]
- 11.Takano K, Hachiya A, Murase D, et al. (2020). Quantitative changes in the secretion of exosomes from keratinocytes homeostatically regulate skin pigmentation in a paracrine manner. J Dermatol, 47:265–276. [DOI] [PubMed] [Google Scholar]
- 12.Jin S, Chen L, Xu Z, Xing X, Zhang C, Xiang L. (2020). 585 nm light-emitting diodes inhibit melanogenesis through upregulating H19/miR-675 axis in LEDs-irradiated keratinocytes by paracrine effect. J Dermatol Sci, 98:102–108. [DOI] [PubMed] [Google Scholar]
- 13.Tulkens J, De Wever O, Hendrix A. (2020). Analyzing bacterial extracellular vesicles in human body fluids by orthogonal biophysical separation and biochemical characterization. Nat Protoc, 15:40–67. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Heikkinen L, Wang C, Yang Y, Sun H, Wong G. (2019). Trends in the development of miRNA bioinformatics tools. Brief Bioinform, 20:1836–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguennouz M, Guarneri F, Oteri R, Polito F, Giuffrida R, Cannavò SP. (2021). Serum levels of miRNA-21-5p in vitiligo patients and effects of miRNA-21-5p on SOX5, beta-catenin, CDK2 and MITF protein expression in normal human melanocytes. J Dermatol Sci, 101:22–29. [DOI] [PubMed] [Google Scholar]
- 16.Shang Z, Li H. (2017). Altered expression of four miRNA (miR-1238-3p, miR-202-3p, miR-630 and miR-766-3p) and their potential targets in peripheral blood from vitiligo patients. J Dermatol, 44:1138–1144. [DOI] [PubMed] [Google Scholar]
- 17.Hushcha Y, Blo I, Oton-Gonzalez L, Mauro GD, Martini F, Tognon M, Mattei M. (2021). microRNAs in the Regulation of Melanogenesis. Int J Mol Sci, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huo J, Liu T, Li F, Song X, Hou X. (2021). MicroRNA-21-5p protects melanocytes via targeting STAT3 and modulating Treg/Teff balance to alleviate vitiligo. Mol Med Rep, 23 (1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seneschal J, Boniface K, D’Arino A, Picardo M. (2021). An update on Vitiligo pathogenesis. Pigment Cell Melanoma Res, 34:236–243. [DOI] [PubMed] [Google Scholar]
- 20.Le L, Sirés-Campos J, Raposo G, Delevoye C, Marks MS. (2021). Melanosome Biogenesis in the Pigmentation of Mammalian Skin. Integr Comp Biol, 61:1517–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu C, Aisa HA. (2017). Upregulation of Melanogenesis and Tyrosinase Activity: Potential Agents for Vitiligo. Molecules, 22(8):1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Yu R, Guo X, et al. (2021). Identification of TYR, TYRP1, DCT and LARP7 as related biomarkers and immune infiltration characteristics of vitiligo via comprehensive strategies. Bioengineered, 12:2214–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L, Wei Y, Sun Y, et al. (2015). Interferon-gamma Inhibits Melanogenesis and Induces Apoptosis in Melanocytes: A Pivotal Role of CD8+ Cytotoxic T Lymphocytes in Vitiligo. Acta Derm Venereol, 95:664–70. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Xue L, Gao H, et al. (2019). Exosomal miRNA derived from keratinocytes regulates pigmentation in melanocytes. J Dermatol Sci, 93:159–167. [DOI] [PubMed] [Google Scholar]
- 25.Wang XY, Guan XH, Yu ZP, et al. (2021). Human amniotic stem cells-derived exosmal miR-181a-5p and miR-199a inhibit melanogenesis and promote melanosome degradation in skin hyperpigmentation, respectively. Stem Cell Res Ther, 12:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Yang X, Liu O, et al. (2021). Differentially expressed microRNAs in peripheral blood mononuclear cells of nonsegmental vitiligo and their clinical significance. J Clin Lab Anal, 35:e23648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Cheng F, He R, Chen H, Liu Y, Sun J. (2014). Inhibition of SATB1 by shRNA suppresses the proliferation of cutaneous malignant melanoma. Cancer Biother Radiopharm, 29:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]