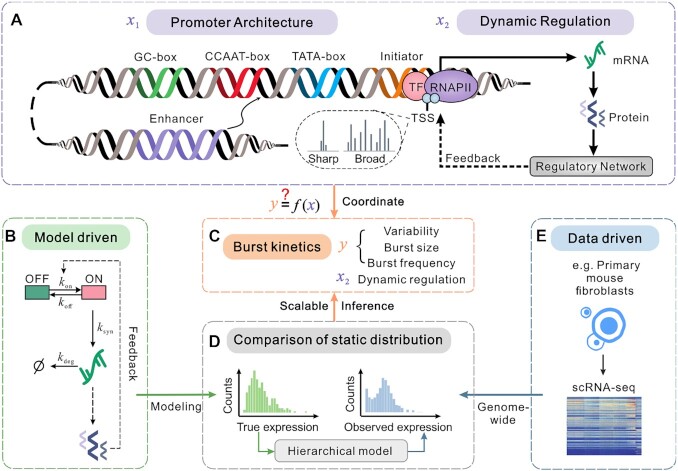
Figure 1.
Overview of a scalable genome-wide inference method. (A) Schematic for important ingredients in gene expression process, including static promoter architecture information and dynamic regulation. Promoter architecture (represented by 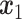 ) consists of promoter motifs (Initiator, TATA-box, CCAAT-box, and GC-box), TSS distributions (‘sharp’ and ‘broad’) and enhancer–promoter interactions. Dynamic regulation (represented by
) consists of promoter motifs (Initiator, TATA-box, CCAAT-box, and GC-box), TSS distributions (‘sharp’ and ‘broad’) and enhancer–promoter interactions. Dynamic regulation (represented by 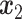 ) is referred to as a series of processes, such as transcription and translation as well as feedback loops, in which the gene product (as a transcription factor) regulates its own expression, possibly via a complex regulatory network. (B) Model-driven: schematic for a mechanistic model of stochastic gene expression, which considers an active (ON) state and an inactive (OFF) state of the promoter and auto-regulatory feedback. Here
) is referred to as a series of processes, such as transcription and translation as well as feedback loops, in which the gene product (as a transcription factor) regulates its own expression, possibly via a complex regulatory network. (B) Model-driven: schematic for a mechanistic model of stochastic gene expression, which considers an active (ON) state and an inactive (OFF) state of the promoter and auto-regulatory feedback. Here 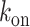 is the switching rate from OFF to ON and
is the switching rate from OFF to ON and 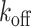 from ON to OFF;
from ON to OFF; 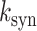 is the transcription rate when the gene is in ON state and
is the transcription rate when the gene is in ON state and 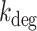 is the degradation rate of mRNAs. (C) Kinetic parameters to be inferred, which include expression variability, burst frequency, burst size, and dynamic regulation. (D) Comparison between two static distributions (the left panel is for ‘true’ mRNA levels in the mechanistic gene model and the right panel for ‘observed’ mRNA counts in a given set of scRNA-seq data) by a hierarchical model can determine the values of the kinetic parameters in (C) via a scalable genome-wide inference method. (E) Data-driven: genome-wide scRNA-seq data of mouse embryonic fibroblasts gives an expression matrix that further gives the observed static distribution in (D).
is the degradation rate of mRNAs. (C) Kinetic parameters to be inferred, which include expression variability, burst frequency, burst size, and dynamic regulation. (D) Comparison between two static distributions (the left panel is for ‘true’ mRNA levels in the mechanistic gene model and the right panel for ‘observed’ mRNA counts in a given set of scRNA-seq data) by a hierarchical model can determine the values of the kinetic parameters in (C) via a scalable genome-wide inference method. (E) Data-driven: genome-wide scRNA-seq data of mouse embryonic fibroblasts gives an expression matrix that further gives the observed static distribution in (D).