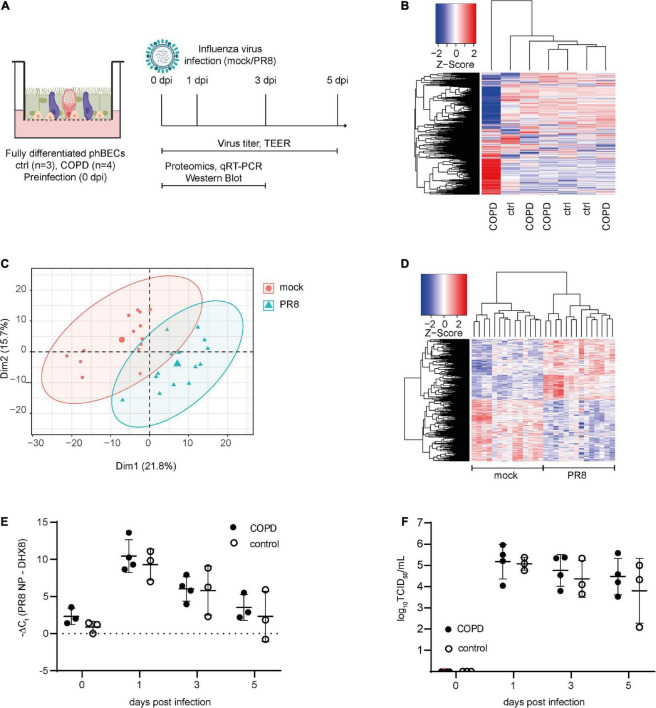
FIGURE 2.
Proteomics pre- and post-influenza virus infection in fully differentiated primary human bronchial epithelial cells (phBECs). PhBECs from patients with chronic obstructive pulmonary disease (COPD) (n = 4) and controls (n = 3) were differentiated for 28 days at the air–liquid interface and either mock-infected or infected with influenza A virus PR8. Cells were harvested for proteomics on day 28 after airlift (pre-infection), and on days 1 and 3 post-infection. To examine viral replication, cells and apical washes were harvested on day 0 (after incubating the cells with the virus to allow attachment, and after removing the unattached virus by washing) and on days 1, 3, and 5 post-infection. (A) Scheme of culture, infection, and sample analysis. An illustration was created with biorender.com. TEER, transepithelial electrical resistance. (B) Heatmap of proteomics pre-infection. The protein profile of the COPD GOLD stage IV-derived phBECs is shown in the outermost left lane. (C) Principal component analysis of proteomics post-infection. (D) Heatmap of significantly altered proteins post-infection with all patients/controls and all time points (PR8 vs. mock, n = 7). A protein was considered to be differentially expressed if the comparison resulted in a false discovery rate (FDR) of less than 5% with the Benjamini–Hochberg (BH) correction to correct multiple testing. (E) Total RNA was extracted from cells and mRNA levels of PR8 nucleoprotein transcripts were examined by RT-qPCR. DHX8 was used as a housekeeping gene. (F) Infectious viral particles in apical washes were quantified by an end point dilution assay. TCID50: 50% tissue culture infectious dose. For panels (E,F), statistical analysis was performed using an unpaired, two-sided Mann–Whitney U test, but with a cutoff point of p < 0.05, no statistically significant differences were observed.