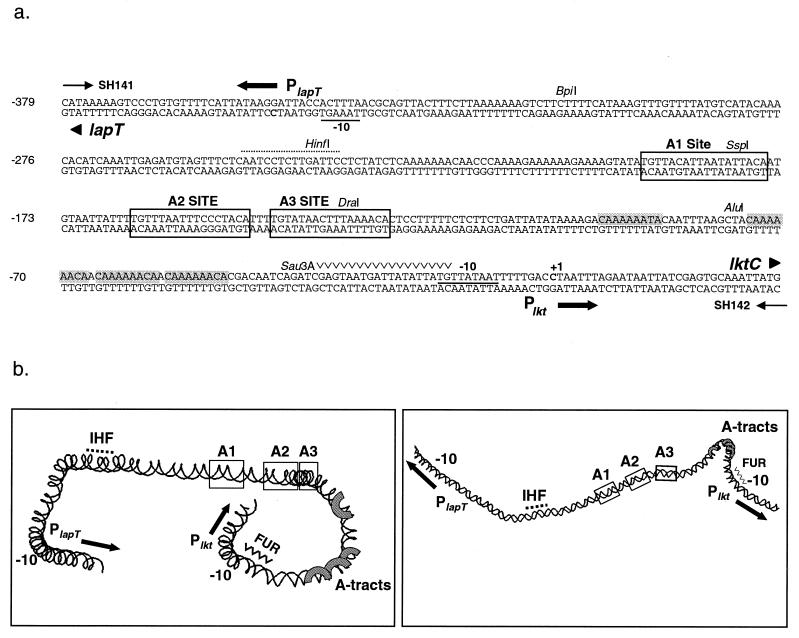
FIG. 1.
(a) Nucleotide sequence of the lapT-lktC intergenic regulatory region of M. haemolytica SH1217. Bolded cytosine residues indicate the transcriptional start sites of PlapT and Plkt. The sequence is numbered in reference to the Plkt transcriptional start site at +1. Underlined nucleotides indicate the predicted −10 promoter regions of PlapT and Plkt. Shaded nucleotides correspond to the four phased adenine tracts. Putative upstream activator sites A1, A2, and A3 are boxed. The near-consensus IHF binding site is indicated with a dotted line above the sequence, and a potential Fur binding site is marked above the nucleotides (∨∨∨∨∨). Unique enzyme restriction sites are shown. (b) Computer-predicted DNA structure of the 0.4-kb lapT/lktC promoter region that was generated by the CURVATURE program (42). Two different views were created by rotating the image along its x axis by using RasMac v2.6 (37). The image on the right has been reduced in size. Sequence elements are noted on the model. IHF and FUR are potential IHF and FUR binding sites, and A-tracts are adenine tracts. Solid arrows show the direction of transcription.