Abstract
Deciphering the genomic profiles and tumor microenvironment (TME) in large cell carcinomas of the lung (LCC), large cell neuroendocrine of the lung (LCNEC), and small cell lung cancer (SCLC) might contribute to a better understanding of lung cancer and then improve outcomes. Ten LCC patients, 12 LCNEC patients, and 18 SCLC patients were enrolled. Targeted next-generation sequencing was used to investigate the genomic profiles of LCC, LCNEC, and SCLC. Tumor-infiltrating lymphocytes (TILs) within cancer cell nests and in cancer stroma were counted separately. Precise 60% of LCNEC patients harbored classical non-small cell lung cancer driver alterations, occurring in BRAF, KRAS, ROS1, and RET. More than 70% of SCLC patients harbored TP53-RB1 co-alterations. Moreover, 88.9%, 40%, and 77.8% of LCC, LCNEC, and SCLC cases had a high tumor mutation burden level with more than 7 mutations/Mb. Furthermore, high index of CD68+ CD163+ (TILs within cancer cell nests/ TILs within cancer cell nests and in cancer stroma, P = .041, 548 days vs not reached) and CD163+ TILs (P = .041, 548 days vs not reached) predicted a shorter OS in SCLC. Our findings revealed the distinct genomic profiles and TME contexture among LCC, LCNEC, and SCLC. Our findings suggest that stratifying LCNEC/SCLC patients based on TME contexture might help clinical disease management.
Keywords: genomic profiles, large cell carcinoma, large cell neuroendocrine, small cell lung cancer, tumor microenvironment
1. Introduction
Lung cancer is one of the most commonly diagnosed cancer and is the leading cause of cancer-related death worldwide,[1] which is histologically classified into adenocarcinoma (ADC), squamous cell carcinoma (SCC), large cell carcinoma (LCC), neuroendocrine tumors, and not otherwise specified tumors. Unlike ADC or SCC, LCC as a rare histological subtype lacks targeted therapies, and specific molecular markers.[2]
Pulmonary neuroendocrine tumors account for about 20% of all primary lung tumors,[3] which consist of preinvasive lesions, neuroendocrine tumors (typical and atypical carcinoids), and neuroendocrine carcinomas (large cell neuroendocrine carcinoma [LCNEC] and small cell carcinoma [SCLC]) according to the 2021 World Health Organization classification.[4,5] Due to the rarity of LCC and LCNEC, parallel comparison of molecular features across these 3 histological subtypes is limited to be documented, especially in eastern Asian populations.
Tumor microenvironment (TME) presents a coordinated network of interface cell types mainly include immune cells, endothelial cells, fibroblasts through the extracellular matrix, cytokines, chemokines, and various metabolites.[5,6] Previous studies have revealed that immune contexture in the TME is related to survival outcomes of lung cancer patients, such as the type, density, and location of tumor-infiltrating lymphocytes (TILs). Osamu et al have reported that CD4+ T cells in cancer stroma are associated with favorable prognosis of non-small cell lung cancer (NSCLC).[7] The density of CD8+ T cells in stroma is an independent prognostic factor associated with disease-free survival, disease-specific survival, and overall survival (OS) of NSCLC.[8] Over the last decade, the advance of immunotherapy, such as immune checkpoint inhibitors (ICIs), have revolutionized the treatment of multiple solid tumor types.[9] ICIs exert their roles by blocking the interaction between molecules involved in the regulation of T cell activation or function, such as the interaction between programmed cell death protein 1 (PD-1), and programmed cell death ligand-1 (PD-L1).[10] Anti-PD-1/anti-PD-L1 agents have been established as standard treatment options in both NSCLC and SCLC. National Comprehensive Cancer Network SCLC guidelines (version 2. 2022) recommend nivolumab or pembrolizumab as first-line treatment for extensive stage SCLC and as the subsequent systemic therapy for relapsed or primary progressive SCLC.[11] High tumor mutational burden (TMB) has been documented in LCNEC, offering treatment options of ICIs.[12] A recent real-world study has revealed an overall survival advantage in advanced LCNEC patients who received ICIs.[13] Although the treatment efficacies of ICIs in LCC have not been well established, several cases have documented the efficacies of ICIs in refractory or metastatic LCC patients.[14–16] However, it is currently recognized that one of the toughest barriers to anti-tumor immunotherapy is immunosuppression created by the tumor microenvironment.[17] There is a need to understand the TME contexture to discover biomarkers that may predict treatment outcomes and prognosis.
In the present work, we performed comprehensive genomic profiling and TME analysis of LCC, LCNEC, and SCLC. The molecular profiles were compared among LCC, LCNEC, and SCLC.
2. Materials and methods
2.1. Patients
Between February 2017 and October 2020, lung cancer patients who met the following inclusion criteria were respectively enrolled: Patients were diagnosed as LCC, LCNEC, and SCLC according to the World Health Organization histological classification of lung tumors[18,19]; Patients were older than 18; Patients underwent surgical resection in Sichuan Provincial People Hospital. A total of 40 patients enrolled, including 10 patients with LCC, 12 with LCNEC, and 18 with SCLC. Pathological or clinical staging was based on the eighth edition of the American Joint Committee on Cancer.[20] All tumors were reviewed by 2 independent pathologists. This study was approved by the Ethics Committee of Sichuan Provincial People Hospital. Informed consent was obtained from patients for the use of their tissue samples. OS was defined as the interval from the date of surgery to the date of death. Patients without an event were censored at the time of the last follow-up visit.
2.2. DNA extraction and DNA library construction
Tissue DNA was extracted with a QIAamp DNA formalin-fixed paraffin-embedded (FFPE) tissue kit (Qiagen, Valencia, CA) according to the manufacturer instructions. The concentration of DNA extracted from tumor tissue was measured by Qubit 2.0 Fluorometer with Qubit double-stranded DNA assay kit (Life Technologies, Carlsbad, CA). A minimum of 50 ng of DNA was used for NGS library preparation. DNA was fragmented by Covaris M220 focused ultrasonicator (Covaris, Inc., Woburn, MA) followed by end repair, phosphorylation, dA addition, and adaptor ligation for library construction. Then, DNA library was purified by using Agencourt AMPure beads (Beckman Coulter, Fullerton, CA).
2.3. Capture-based targeted sequencing
Capture-based targeted sequencing was performed on samples in a Clinical Laboratory Improvement Amendments-certified, College of American Pathology (CAP)-accredited laboratory using a panel consisting of 520 cancer-related genes spanning 1.64 megabases (Mb) of the human genome (OncoScreenPlus, Burning Rock Biotech, Guangzhou, China).[21] Indexed samples were sequenced on Nextseq500 (Illumina, Inc., San Diego, CA) with paired-end reads and a mean sequencing depth of 1698×.
2.4. Sequencing data analysis
Sequencing data is processed using a customized bioinformatics pipeline designed to detect several classes of genomic alterations, including single-nucleotide variants, small insertions, and deletions, large genomic rearrangements (LGRs), copy number variants (CNV), and genomic fusions. The raw sequencing data were preprocessed by using bcl2fastq. Preprocessed sequencing data were mapped to the human genome (hg19) by using Burrows-Wheeler Aligner 0.7.10 to generate BAM files. Variant calling was performed by using Vardict to detect single-nucleotide variants/insertions and deletions. Variants were annotated with ANNOVAR and SnpEff v3.6. The maximum allele frequencies of genomic alterations were calculated. Factera was used to identify genomic fusions and calculate their abundance. The CNV and LGR were estimated with an in-house algorithm based on the sequencing depth as described previously,[22,23] respectively. TMB per patient was computed as a ratio between the total number of nonsynonymous mutations detected with the coding region size of the panel using the formula below. CNV, fusions, LGRs, and mutations occurring on the kinase domains on EGFR and ALK were excluded from the mutation count.[21]
2.5. Multiplex immunofluorescence (mIF) assays
mIF assays were performed for serial FFPE slides to visualize TILs using PANO 7-plex IHC kit (Panovue, Beijing, China). Primary antibodies against cell differentiation (CD)3, CD8, CD56, CD68, CD163, PD-1, and PD-L1 were sequentially applied, followed by horseradish peroxidase-conjugated secondary antibody incubation and tyramide signal amplification. Nuclei were stained with 4′-6′-diamidino-2-phenylindole (DAPI) after all the antigens had been labeled. Two serial FFPE sections form the same tissue sample were performed for mIF analysis. One section was stained with CD3 (indigo), CD8 (red), PD-1 (green), PD-L1 (yellow), panCK (purple), and DAPI (blue). Another section was stained with CD56 (yellow), CD163 (red), CD68 (green), panCK (purple), and DAPI (blue). The stained slides were scanned using the Mantra System (PerkinElmer, Waltham, Massachusetts), which captures the fluorescent spectra at 20-nm wavelength intervals from 420 to 720 nm. Images analyses were performed using inform image analysis software (PerkinElmer, Waltham, Massachusetts).[24] TILs within cancer cell nests (iTILs) and in cancer stroma (sTILs) were counted separately.
2.6. Statistical analysis
Differences in 2-groups were accessed by Chi-square test or by Fisher exact test for categorical variables. P value < 0.05 was considered statistically significant. The median density of a certain iTIL/sTIL was used as the cutoff to divide patients into 2 groups, respectively. The correlations of TILs with survival outcomes were analyzed using Kaplan–Meier curves and Log-rank tests. All statistical analyses were performed in R 4.0.5 (https://cran.r-project.org/src/base/R-4/) as a free software environment for statistical computing and graphics (The R Project for Statistical Computing, https://www.r-project.org/).
3. Results
3.1. Clinicopathological characteristics
We performed a comprehensive genomic profiling spanning 3 subtypes of lung tumors in the Chinese population. A total of 40 patients were enrolled, including 14 patients with stage I, 10 patients with stage II, 12 patients with stage III, 1 patient with stage IV, and the remaining 3 patients with unknown tumor stage. The cohort had 34 (85%) males and a median age of 63 years (ranging: 46–80 years). Of which, 36 patients at stage I-IIIA underwent radical resection and 1 patient at stage IV underwent palliative resection. The epidemiological characteristics of patients with different histological subtypes are summarized in Table S1, Supplemental Digital Content, http://links.lww.com/MD/I383 (Supplemental Digital Content, which illustrates the clinicopathological characteristics of patients), which describes the similar clinicopathological characteristics among LCC, LCNEC, and SCLC in age, gender distribution, and Ki-67 expression.
3.2. Genomic profiles of LCC, LCNEC, and SCLC
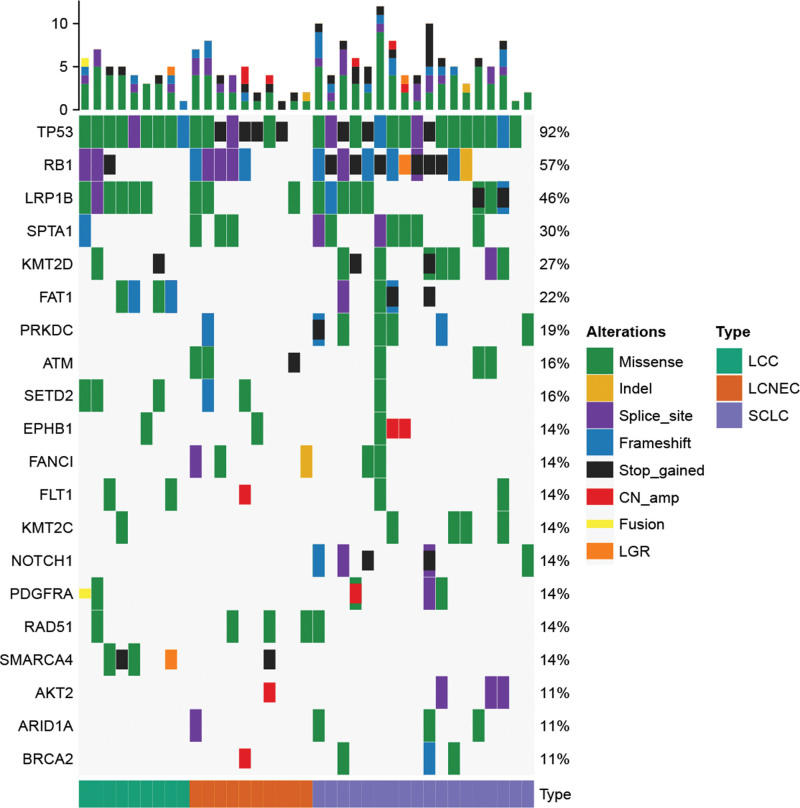
In this work, 37 patients (9 with LCC, 10 with LCNEC, and 18 with SCLC) who had available archived tissue samples underwent capture-based targeted sequencing. A total of 551 somatic alterations occurring in 219 genes were identified. Each patient harbored at least 1 somatic alteration detected from this panel. The most frequently mutated genes were TP53 (92%), RB1 (57%), and LRP1B (46%) (Fig. 1). TP53 was the most frequently altered gene in LCC (100%, 10/10), LCNEC (83.3%, 10/12), and SCLC (94.4%, 17/18), respectively. LRP1B alterations occurred in 60.0% (6/10) of LCC patients. RB1 alterations occurred in 41.7% (5/12) of LCNEC patients. RB1, LRP1B, and KMT2D alterations were observed in 77.8% (14/18), 61.1% (11/18), and 50.0% (9/18) of SCLC patients. SCLC harbored more TP53-RB1 co-alterations than LCC. SMARCA4 and SETD2 were altered in 44.4% and 33.3% of LCC patients, while SMARCA4 or SETD2 alterations were not observed in SCLC.
Figure 1.
The comparison of genomic features in different histological sub-cohorts. The oncoprint of genomic alterations identified in LCC, LCNEC, and SCLC. LCC = large cell carcinoma, LCNEC = large cell neuroendocrine carcinoma, SCLC = small cell lung cancer.
3.3. Tumor mutational burden of patients with LCC, LCNEC, and SCLC
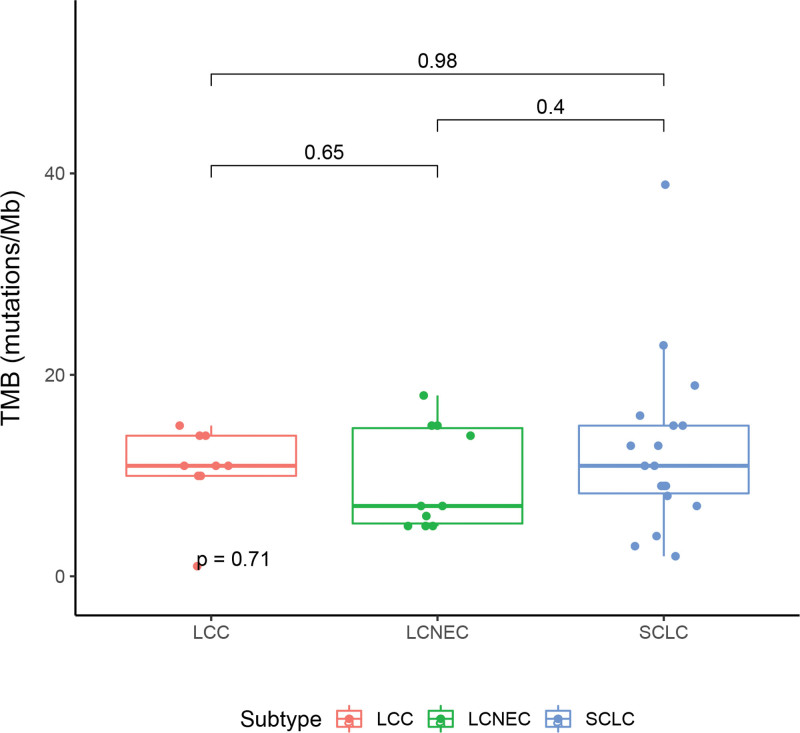
Next, we estimated the TMB of patients with LCC, LCNEC, and SCLC. TMB was calculated as described in the “Methods” section. Nine, 10, and 18 patients with LCC, LCNEC, and SCLC had available TMB, respectively. The median TMB of patients with LCC was 10.97 mutations/Mb (ranging from 1 to 14.96), which was 6.98 (ranging from 4.99 to 17.95) and 10.97 mutations/Mb (ranging from 1.99 to 38.88) in LCNEC and SCLC, respectively (Fig. 2). Taken together, we observed a comparable TMB levle among LCC, LCNEC, and SCLC. We also found that 88.9% (8/9), 40.0% (4/10), and 77.8% (14/18) of LCC, LCNEC, SCLC cases had a high TMB level with more than 7 mutations/Mb.
Figure 2.
The comparison of TMB level in different histological sub-cohorts. TMB = tumor mutation burden.
3.4. Tumor microenvironment in patients with LCC, LCNEC, and SCLC
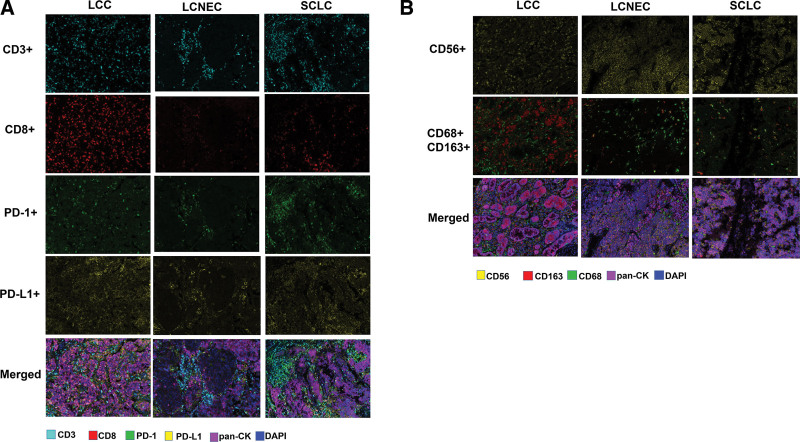
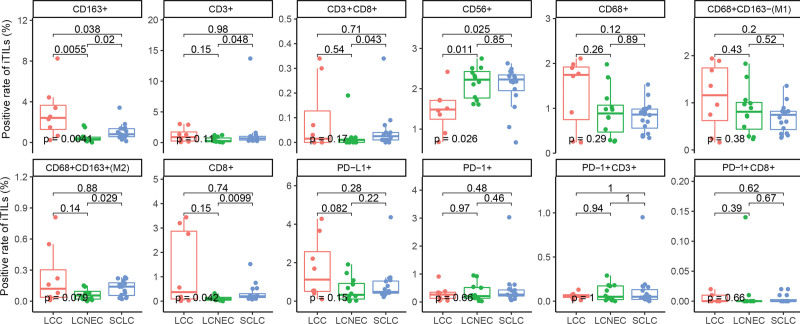
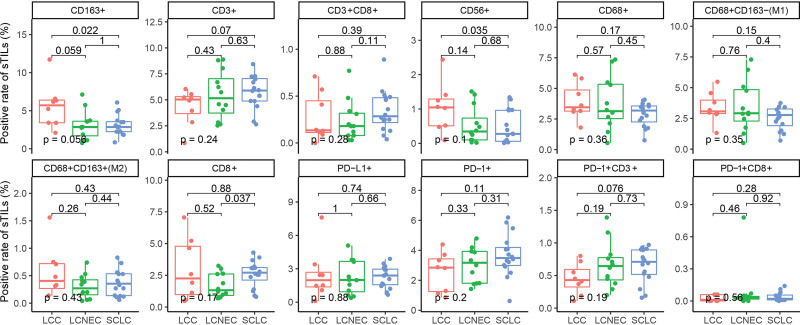
We also assessed the tumor microenvironment in patients with LCC, LCNEC, and SCLC. A total of 36 patients who had sufficient archived tissue samples underwent mIF assays to visualize TILs, including 8 LCCs, 12 LCNECs, and 16 SCLCs. The representative mIF images of surgical resection samples analyzed for immune-related biomarkers are shown in Figure 3A–3B. In the study, SCLC displayed a significantly higher rate of CD3+ iTILs (Fig. 4, P = .048), CD8+ iTILs (Fig. 4, P = .0099), CD3+ CD8+ iTILs (Fig. 4, P = .043), and CD68+ CD163+ iTILs (Fig. 4, P = .029) in comparation with LCENC. Moreover, LCC had a significantly higher rate of CD163+ iTILs than LCENC (Fig. 4, P = .0055) and SCLC (Fig. 4, P = .038), respectively. The difference of rate of sTILs among LCC, LCNEC, and SCLC was also investigated. SCLC had a significantly lower rate of CD163+ sTILs compared with LCC (Fig. 5, P = .022) and showed higher rate of CD8+ sTILs than LCNEC (Fig. 5, P = .037).
Figure 3.
Representative multiplex immunofluorescence images of tissue samples obtained from LCC, LCENC, and SCLC. (A) Representative images regarding mIF staining for CD3, CD8, PD-1, and PD-L1; (B) representative images regarding mIF staining for CD56, CD68, and CD163. DAPI = 4’,6-diamidino-2-phenylindole, LCC = large cell carcinoma, LCNEC = large cell neuroendocrine carcinoma, mIF = multiplex immunofluorescence, pan-CK = pan-cytokeratin, PD-1 = programmed cell death, PD-L1 = programmed cell death ligand-1, SCLC = small cell lung cancer.
Figure 4.
The comparison of iTILs in different histological sub-cohorts. iTILs = TILs within cancer cell nest.
Figure 5.
The comparison of sTILs in different histological sub-cohorts. sTILs = TILs in cancer stroma.
3.5. The association of TILs with survival outcome in patients with LCNEC and SCLC
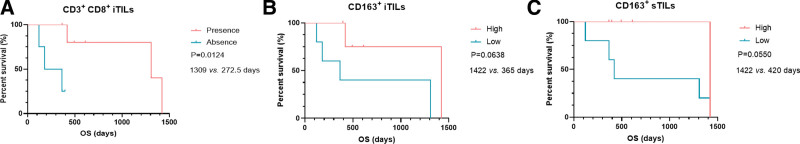
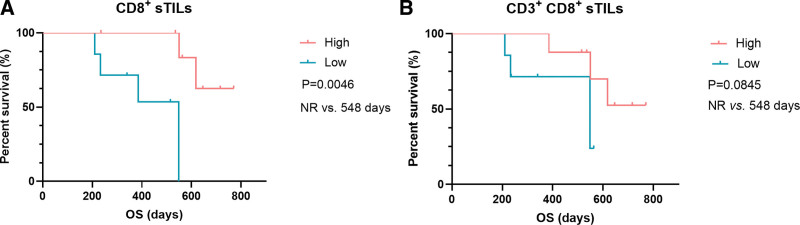
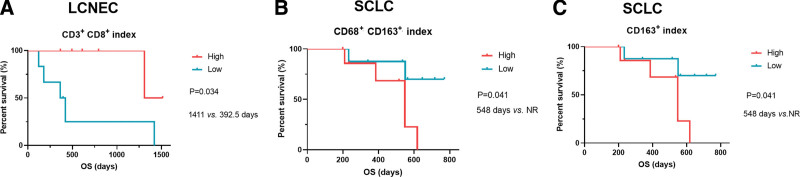
Given that all LCC patients were still alive at the end of follow-up, the associations between iTILs/sTILs, and survival outcomes were explored in patients with LCNEC (n = 10) and SCLC (n = 14), respectively. We found LCNEC patients with the presence of CD3+CD8+ iTILs (n = 6) exhibited a significantly longer OS than those with the absence of CD3+CD8+ iTILs (n = 4) (median OS [mOS]: 1309 vs 272.5 days, P = .0124, Fig. 6A, Table S2, Supplemental Digital Content, http://links.lww.com/MD/I384 [Supplemental Digital Content, which illustrates the survival of patients in different groups]). We also found that LCNEC patients with high level of CD163+ iTILs and CD163+ sTILs had a trend of longer OS than those with low level of CD163+ iTILs (mOS: 1422 vs 365 days, P = .0638, Fig. 6B, Table S2, Supplemental Digital Content, http://links.lww.com/MD/I384 [Supplemental Digital Content, which illustrates the survival of patients in different groups]) and CD163+ sTILs (mOS: 1422 vs 420 days, P = .0550, Fig. 6C, Table S2, Supplemental Digital Content, http://links.lww.com/MD/I384 [Supplemental Digital Content, which illustrates the survival of patients in different groups]), respectively. In SCLC, high level of CD8 + sTILs was significantly associated with a favorable OS (mOS: not reached [NR] vs 548 days, P = .0046, Fig. 7A, Table S1, Supplemental Digital Content, http://links.lww.com/MD/I384 [Supplemental Digital Content, which illustrates the survival of patients in different groups]). Moreover, a trend of significantly longer OS was observed in patients with a high level of CD3+CD8+ sTILs (mOS: NR vs 548 days, P = .0845, Fig. 7B, Table S2, Supplemental Digital Content, http://links.lww.com/MD/I384 [Supplemental Digital Content, which illustrates the survival of patients in different groups]). In addition, the associations between iTILs/sTILs index (the rate of iTILs/the rate of sTILs) and OS were also explored in LCNEC and SCLC. We found that high index of CD3+CD8+ TILs predicted a longer OS in LCNEC (mOS: 1141 vs 392.5 days, P = .034, Fig. 8A, Table S2, Supplemental Digital Content, http://links.lww.com/MD/I384 [Supplemental Digital Content, which illustrates the survival of patients in different groups]). Moreover, high index of CD68+CD163+ (Fig. 8B, P = .041, mOS: 548 days vs NR, Table S2, Supplemental Digital Content, http://links.lww.com/MD/I384 [Supplemental Digital Content, which illustrates the survival of patients in different groups]) and CD163+ TILs (mOS: 548 days vs NR, P = .041, Fig. 8C, Table S2, Supplemental Digital Content, http://links.lww.com/MD/I384 [Supplemental Digital Content, which illustrates the survival of patients in different groups]) predicted a shorter OS in SCLC.
Figure 6.
The associations of TILs with OS in LCNEC. (A). The association of CD3+ CD8+ iTILs with OS; (B). the association of CD163+ iTILs with OS; (C). the association of CD163+ sTILs with OS. CD = cell differentiation, iTILs = TILs within cancer cell nests, LCNEC = large cell neuroendocrine carcinoma, OS = overall survival, sTILs = TILs in cancer stroma, TILs = tumor-infiltrating lymphocytes.
Figure 7.
The associations of TILs with OS in SCLC. (A) The association of CD8+ sTILs with OS; (B) the association of CD3+ CD8+ sTILs with OS. CD = cell differentiation, iTILs = TILs within cancer cell nests, OS = overall survival, SCLC = small cell lung cancer, sTILs = TILs in cancer stroma, TILs = tumor-infiltrating lymphocytes.
Figure 8.
The associations between iTILs/sTILs index and OS in LCNEC, and SCLC. iTILs = TILs within cancer cell nests, iTILs/sTILs index = the rate of iTILs/the rate of sTILs, LCNEC = large cell neuroendocrine carcinoma, OS = overall survival, sTILs = TILs in cancer stroma, SCLC = small cell lung cancer.
4. Discussion
In the present work, we found that TP53 was the most frequently mutated gene in LCC, which was consistent with the previous study.[25] TP53 was also identified as the most commonly mutated gene in LCNEC and SCLC in previous and our studies.[26–28] It is well known that SCLC is characterized by TP53/RB1 co-mutation/loss that about 80% of cases harbor concurrent TP53 and RB1 alterations.[28] Similar results were also observed in this study that 72.3% of SCLC patients harbored TP53-RB1 co-alterations. Rekhtman et al[26] have classified LCNEC into an SCLC-like subset characterized by TP53/RB1 co-mutation/loss, and an NSCLC-like subset characterized by the lack of TP53/RB1 co-alteration but the presence of NSCLC-type mutations. TP53-RB1 co-alteration was observed in 50.0% of Chinese LCNEC patients, reproducing the results from the previous studies.[26,27,29] We observed 60.0% of LCNEC cases harboring classical NSCLC driver gene alterations, including 2 missense mutations in BRAF (p.G466E and p.N580I), 1 KRAS amplification, 1 KRAS missense mutation p.G12A, 1 ROS1 missense mutation p.V1002A, and 1 RET missense mutation p.G568S. The mutation rate of classical NSCLC driver genes in this work was higher than that reported by Miyosh et al[30] (60.0% vs 23.0%), which suggests the unique molecular characteristics in Chinese LCNEC.
Chan et al[25] have revealed that TP53 (84.7%, 50/59), KRAS/NRAS/HRAS (20.3%, 12/59), and PIK3CA (16.9%, 10/59) were the most commonly altered genes in LCCs. In this work, TP53 (100%, 9/9) was also identified as the most frequently altered gene, followed by LRP1B (66.7%, 6/9) and FAT1 (44.4%, 4/9). In addition, KRAS/NRAS/HRAS and PIK3CA alterations were not observed in LCC. The difference of genomic profiling in LCC between our and the previous study might be attributed to several factors. First, LCC is a heterogenous group of primary lung cancers that lacks any clear morphological or immunohistochemical differentiation toward small cell carcinoma, ADC, or SCC. Second, the genetic architecture of LCC in Chinese and Western population might be distinct. SMARCA4 (40%, 4/10) was identified as one of most frequently altered genes in LCC. SMARCA4-deficient lung cancer has been considered a separate entity.[31] The prevalence of altered SMARCA4 might be biased by the presence of a high prevalence of this particular genotype. SMARCA4/BRG1 immunohistochemistry would be needed to characterize these cases, to assess if there is loss of the protein product or if the alteration is only heterozygous.
ICIs targeting the PD-1/PD-L1 axis elicit remarkable clinical efficacy in a spectrum of malignancies,[9,32,33] which have emerged as a pillar of standard cancer care. There are no accepted universal biomarkers capable to accurately predict response to ICIs. Of note, TMB has emerged as a potential biomarker for predicting the response to ICIs. Several previous studies have reported that SCLC is characterized by high TMB,[34,35] however, the TMB status in LCC and LCNEC remains elusive. In this study, we found that 88.9% (8/9), 40.0% (4/10), and 77.8% (14/18) of LCC, LCNEC, SCLC cases had a high TMB level with more than 7 mutations/Mb detected from the OncoScreen panel. The previous study has demonstrated that advanced NSCLC patients with TMB ≥7 mutations/Mb detected from the OncoScreen panel exhibited a significantly longer PFS than those with TMB <7 mutations/Mb after receiving ICI treatment in a Chinese population.[21] A comparable TMB status was observed among LCC, LCNEC, and SCLC (median TMB: 10.97 vs 6.98 vs 10.97 mutations/Mb) in this work. It has been documented that the median TMB of Chinese lung ADC patients is 4.61 mutations/Mb, while the median TMB of the Caucasian patients derived from The Cancer Genome Atlas (TCGA) is 3.26 mutations/Mb.[36] The median TMB of Chinese lung SCC is 9.43 mutations/Mb.[37] These findings suggest that LCC, LCNEC, and SCLC might have a high level of TMB than lung ADC in Chinese patients. These findings also raise the potential of utilizing ICIs as treatment regimens in LCC, LCNEC, and SCLC patients who had high TMB level.
CD8+ T cells are the key components of the adaptive immune response, which is considered to be the main effector population in mediating immunity contributing to the clearance of tumor cells.[38,39] In this study, SCLC displayed more infiltration with CD3+ iTILs, CD8+ iTILs, CD8+ sTILs, and CD3+CD8+ iTILs than LCENC. Furthermore, a higher rate of CD8+ iTILs and CD3+CD8+ sTILs predicted a longer OS in SCLC patients, which were consistent with previous studies indicating that a high level of CD8+ lymphocytes is associated with a longer OS.[40,41] Shirasawa et. al[42] have revealed that a low peripheral blood neutrophil to lymphocyte ratio predicted a poor OS in advanced LCNEC patients and NLR was inversely correlated with CD8+ iTILs and CD8+ sTILs. In this study, low rate of CD3+CD8+ iTILs associated with an unfavorable OS in LCNEC patients was observed. These findings indicate that low level of CD8+ iTILs might be a predictor of a poor OS in LCNEC. Further studies are needed to explore the prognostic value of CD8+ iTILs in LCNEC. To the best of our knowledge, this is the first study to reveal the TME status in LCC. While the prognostic roles of TILs in LCC were not further investigated due to a small sample size of LCCs.
Macrophages in the TME are well known to associate with tumor development. Activated macrophages are commonly categorized into M1-like (CD68+CD163-) and M2-like (CD68+CD163+) macrophages.[43] Both M1-like and M2-like macrophage are closely related to inflammatory response, but play distinct roles in tumor development.[44] M1-like macrophages can secrete classic inflammatory cytokines that kill tumors by promoting tumor cell necrosis and immune cell infiltration into the tumor microenvironment.[44,45] In contrast, M2-like macrophages exhibit powerful tumor-promoting functions, including degradation of tumor extracellular matrix, destruction of basement membrane, promotion of angiogenesis, and recruitment of immunosuppressor cells, all of which further promote tumor progression and distal metastasis.[44,45] In this work, LCC showed a higher level of CD163+ iTILs than LCNEC and SCLC. Moreover, LCNEC patients with the presence of CD163+ iTILs had a longer OS. We also found that SCLC had more CD68+ CD163+ iTILs than LCNEC, which was consistent with the previous study indicating that SCLC is enriched in profibrotic and immunosuppressive monocytes/macrophages.[46].
Human natural killer (NK) cells are defined as CD56+CD3-,[47] which are powerful effectors of innate immunity that have significant capability in tumor immune-surveillance as a first line of defense against cancer.[48,49] In this work, we found that the rate of CD56+ iTILs in LCC was significantly lower than that in LCNEC (P = .011) and SCLC (P = .025), respectively. These findings indicated that humoral immune response to the antitumor effects mediated by NK cells might be suppressed in LCC. Further studies are needed to explore the associations of NK cells with LCC tumorigenesis and metastasis.
Gay et al[50] have revealed that SCLC with a neuroendocrine low phenotype and inflamed phenotype having higher expression of immune checkpoint molecules exhibit an improved survival in response to anti-PD-L1, agents combined with platinum-based therapy, compared with other SCLC subtypes. In this work, we found that high level of CD8+ sTILs was significantly associated with a favorable OS in SCLC. These findings highlight the potential utilization of TME contexture in stratifying SCLC or lung cancer patients for disease management.
There are several limitations in this study. First, the sample size was small, which resulted in the bias of our conclusions. Although the prognostic significance of CD3+ CD8+ iTILs, CD163+ iTIL, and CD163+ sTILs in LCNEC, and CD8+ sTILs and CD3+ CD8+ sTILs in SCLC was preliminarily observed based on univariate cox analyses, a large cohort study is needed to explore the prognostic significance of TILs adjusted by adjuvant chemotherapy, type of surgery, age, gender, and disease stage based on multivariate cox analyses. Second, due to the small sample size of LCC patients, the associations of TILs with survival outcomes of LCC patients were not explored in this work. Further studies are needed to investigate the prognostic values of TILs in LCC patients. Third, the associations between alterations and survival in LCC, LCNEC, and SCLC patients should be further explored in a large cohort. In addition, the prognostic value of alterations combined with TME should be further investigated in LCC, LCNEC, and SCLC patients.
5. Conclusions
In conclusion, this is the first study to compare the genomic landscape and TME among LCC, LCNEC, and SCLC. Our data indicate that genomic profiles and TME contexture may complement histological evaluation to provide prognostic and therapeutic stratification, which might help clinical management.
Acknowledgments
We would like to express our gratitude to Haiwei Du, Xuan Lin, and Yufang Wang from Burning Rock Biotech for their valuable assistance in data analysis and interpretation, and manuscript development.
Author contributions
Conceptualization: Dandan Dong.
Data curation: Fanghua Li, Yue Yang, Ying Xu, Ke Li, Linhong Song, Yang Xue.
Formal analysis: Fanghua Li, Ying Xu, Ke Li, Linhong Song, Dandan Dong.
Investigation: Fanghua Li, Linhong Song, Yang Xue.
Methodology: Fanghua Li, Yue Yang, Linhong Song, Yang Xue.
Resources: Fanghua Li, Yue Yang, Yang Xue.
Software: Fanghua Li, Yue Yang, Yang Xue.
Validation: Fanghua Li, Linhong Song.
Visualization: Fanghua Li, Dandan Dong.
Writing – original draft: Fanghua Li, Yue Yang, Linhong Song, Yang Xue, Dandan Dong.
Writing – review & editing: Dandan Dong.
Supplementary Material
Abbreviations:
- ADC
- adenocarcinoma
- CNV
- copy number variants
- DAPI
- 4′-6′-diamidino-2-phenylindole
- FFPE
- formalin-fixed paraffin-embedded
- ICI
- immune checkpoint inhibitor
- iTILs
- TILs within cancer cell nest
- LCC
- large cell carcinoma
- LCNEC
- large cell neuroendocrine carcinoma
- LGR
- large genomic rearrangement
- mIF
- multiplex immunofluorescence
- Mb
- Megabase
- NK
- natural killer
- NR
- not reached
- NSCLC
- non-small cell lung cancer
- OS
- overall survival
- PD-1
- programmed cell death
- PD-L1
- programmed cell death ligand-1
- SCC
- squamous cell carcinoma
- SCLC
- small cell carcinoma
- sTILs
- TILs in cancer stroma
- TILs
- tumor-infiltrating lymphocytes
- TMB
- tumor mutational burden
- TME
- tumor microenvironment
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Supplemental Digital Content is available for this article.
The authors have no funding and conflicts of interest to disclose.
This study was approved by the Ethics Committee of the Sichuan Provincial People’s Hospital.
Informed consent was obtained from each patient for the use of their tumor tissue samples.
How to cite this article: Li F, Yang Y, Xu Y, Li K, Song L, Xue Y, Dong D. Comparative study of the genomic landscape and tumor microenvironment among large cell carcinoma of the lung, large cell neuroendocrine of the lung, and small cell lung cancer. Medicine 2023;102:4(e32781).
Contributor Information
Fanghua Li, Email: 416695981@qq.com.
Yue Yang, Email: 1553007398@qq.com.
Ying Xu, Email: sallyxu@foxmail.com.
Ke Li, Email: 416695981@qq.com.
Linhong Song, Email: songyesong@126.com.
Yang Xue, Email: 1659815999@qq.com.
Reference
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Pelosi G, Barbareschi M, Cavazza A, et al. Large cell carcinoma of the lung: a tumor in search of an author. A clinically oriented critical reappraisal. Lung Cancer. 2015;87:226–31. [DOI] [PubMed] [Google Scholar]
- [3].Rekhtman N. Neuroendocrine tumors of the lung: an update. Arch Pathol Lab Med. 2010;134:1628–38. [DOI] [PubMed] [Google Scholar]
- [4].Metovic J, Barella M, Bianchi F, et al. Morphologic and molecular classification of lung neuroendocrine neoplasms. Virchows Arch. 2021;478:5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Elia I, Haigis MC. Metabolites and the tumour microenvironment: from cellular mechanisms to systemic metabolism. Nat Metab. 2021;3:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Banerjee A, Chabria Y, Kanna NRR, et al. Role of tumor specific niche in colon cancer progression and emerging therapies by targeting tumor microenvironment. Adv Exp Med Biol. 2021;1341:177–92. [DOI] [PubMed] [Google Scholar]
- [7].Wakabayashi O, Yamazaki K, Oizumi S, et al. CD4+ T cells in cancer stroma, not CD8+ T cells in cancer cell nests, are associated with favorable prognosis in human non-small cell lung cancers. Cancer Sci. 2003;94:1003–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Donnem T, Hald SM, Paulsen EE, et al. Stromal CD8+ T-cell density – a promising supplement to TNM staging in non-small cell lung cancer. Clin Cancer Res. 2015;21:2635–43. [DOI] [PubMed] [Google Scholar]
- [9].Bagchi S, Yuan R, Engleman EG. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annu Rev Pathol. 2021;16:223–49. [DOI] [PubMed] [Google Scholar]
- [10].Sadeghi Rad H, Monkman J, Warkiani ME, et al. Understanding the tumor microenvironment for effective immunotherapy. Med Res Rev. 2021;41:1474–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].NCC Network. NCCN Clinical Practice Guidelines in Oncology (NCCN guidelines) Small Cell Lung Cancer (Version 2.2022). Fort Washington: NCCN. 2022. Available at: http://www.nccn.org. [access date November 24, 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yarchoan M, Albacker LA, Hopkins AC, et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight. 2019;4:e126908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dudnik E, Kareff S, Moskovitz M, et al. Real-world survival outcomes with immune checkpoint inhibitors in large-cell neuroendocrine tumors of lung. J ImmunoTher Cancer. 2021;9:e001999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ooi R, Tobino K, Sakabe M, et al. A case of large-cell lung carcinoma successfully treated with pembrolizumab but complicated with cholangitis. Respir Med Case Rep. 2020;31:101197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Okabe N, Mine H, Takagi H, et al. Pulmonary large cell carcinoma, highly positive for PD-L1, shows marked response to pembrolizumab: a case report. Thorac Cancer. 2021;12:1141–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang G, Chai Q, Xiao Y, et al. Case report: therapeutic response to chemo-immunotherapy in an advanced large cell lung carcinoma patient with low values of multiple predictive biomarkers. Front Immunol. 2020;11:607416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Han S, Wang W, Wang S, et al. Tumor microenvironment remodeling and tumor therapy based on M2-like tumor associated macrophage-targeting nano-complexes. Theranostics. 2021;11:2892–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–60. [DOI] [PubMed] [Google Scholar]
- [19].Nicholson AG, Tsao MS, Beasley MB, et al. The 2021 WHO classification of lung tumors: impact of advances since 2015. J Thorac Oncol. 2022;17:362–87. [DOI] [PubMed] [Google Scholar]
- [20].van Roessel S, Kasumova GG, Verheij J, et al. International validation of the eighth edition of the American Joint Committee on Cancer (AJCC) TNM staging system in patients with resected pancreatic cancer. JAMA Surg. 2018;153:e183617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chen X, Fang L, Zhu Y, et al. Blood tumor mutation burden can predict the clinical response to immune checkpoint inhibitors in advanced non-small cell lung cancer patients. Cancer Immunol Immunother. 2021;70:3513–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liu Z, Shi M, Li X, et al. HER2 copy number as predictor of disease-free survival in HER2-positive resectable gastric adenocarcinoma. J Cancer Res Clin Oncol. 2021;147:1315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wu D, Xie Y, Jin C, et al. The landscape of kinase domain duplication in Chinese lung cancer patients. Ann Transl Med. 2020;8:1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stack EC, Wang C, Roman KA, et al. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70:46–58. [DOI] [PubMed] [Google Scholar]
- [25].Chan AW, Chau SL, Tong JH, et al. The landscape of actionable molecular alterations in immunomarker-defined large-cell carcinoma of the lung. J Thorac Oncol. 2019;14:1213–22. [DOI] [PubMed] [Google Scholar]
- [26].Rekhtman N, Pietanza MC, Hellmann MD, et al. Next-generation sequencing of pulmonary large cell neuroendocrine carcinoma reveals small cell carcinoma-like and non-small cell carcinoma-like subsets. Clin Cancer Res. 2016;22:3618–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhuo M, Guan Y, Yang X, et al. The prognostic and therapeutic role of genomic subtyping by sequencing tumor or cell-free DNA in pulmonary large-cell neuroendocrine carcinoma. Clin Cancer Res. 2020;26:892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].George J, Walter V, Peifer M, et al. Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nat Commun. 2018;9:1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Miyoshi T, Umemura S, Matsumura Y, et al. Genomic profiling of large-cell neuroendocrine carcinoma of the lung. Clin Cancer Res. 2017;23:757–65. [DOI] [PubMed] [Google Scholar]
- [31].Chatzopoulos K, Boland JM. Update on genetically defined lung neoplasms: NUT carcinoma and thoracic SMARCA4-deficient undifferentiated tumors. Virchows Arch. 2021;478:21–30. [DOI] [PubMed] [Google Scholar]
- [32].Carlino MS, Larkin J, Long GV. Immune checkpoint inhibitors in melanoma. Lancet. 2021;398:1002–14. [DOI] [PubMed] [Google Scholar]
- [33].Marin-Acevedo JA, Kimbrough EO, Lou Y. Next generation of immune checkpoint inhibitors and beyond. J Hematol Oncol. 2021;14:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hellmann MD, Callahan MK, Awad MM, et al. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell. 2018;33:853–61.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Peifer M, Fernández-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gu W, Wang N, Gu W, et al. Molecular gene mutation profiles, TMB and the impact of prognosis in Caucasians and east Asian patients with lung adenocarcinoma. Transl Lung Cancer Res. 2020;9:629–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jiang T, Shi J, Dong Z, et al. Genomic landscape and its correlations with tumor mutational burden, PD-L1 expression, and immune cells infiltration in Chinese lung squamous cell carcinoma. J Hematol Oncol. 2019;12:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hashimoto M, Kamphorst AO, Im SJ, et al. CD8 T cell exhaustion in chronic infection and cancer: opportunities for interventions. Annu Rev Med. 2018;69:301–18. [DOI] [PubMed] [Google Scholar]
- [39].Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu Rev Immunol. 2007;25:171–92. [DOI] [PubMed] [Google Scholar]
- [40].Eerola AK, Soini Y, Pääkkö P. A high number of tumor-infiltrating lymphocytes are associated with a small tumor size, low tumor stage, and a favorable prognosis in operated small cell lung carcinoma. Clin Cancer Res. 2000;6:1875–81. [PubMed] [Google Scholar]
- [41].Sun Y, Zhai C, Chen X, et al. Characterization of PD-L1 protein expression and CD8(+) tumor-infiltrating lymphocyte density, and their associations with clinical outcome in small-cell lung cancer. Transl Lung Cancer Res. 2019;8:748–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shirasawa M, Yoshida T, Horinouchi H, et al. Prognostic impact of peripheral blood neutrophil to lymphocyte ratio in advanced-stage pulmonary large cell neuroendocrine carcinoma and its association with the immune-related tumour microenvironment. Br J Cancer. 2021;124:925–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yunna C, Mengru H, Lei W, et al. Macrophage M1/M2 polarization. Eur J Pharmacol. 2020;877:173090. [DOI] [PubMed] [Google Scholar]
- [44].Cheng N, Bai X, Shu Y, et al. Targeting tumor-associated macrophages as an antitumor strategy. Biochem Pharmacol. 2021;183:114354. [DOI] [PubMed] [Google Scholar]
- [45].Pan Y, Yu Y, Wang X, et al. Tumor-associated macrophages in tumor immunity. Front Immunol. 2020;11:583084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chan JM, Quintanal-Villalonga A, Gao VR, et al. Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell. 2021;39:1479–1496.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gunesch JT, Dixon AL, Ebrahim TA, et al. CD56 regulates human NK cell cytotoxicity through Pyk2. Elife. 2020;9:e57346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Valipour B, Velaei K, Abedelahi A, et al. NK cells: an attractive candidate for cancer therapy. J Cell Physiol. 2019;234:19352–65. [DOI] [PubMed] [Google Scholar]
- [49].Guillerey C. NK cells in the tumor microenvironment. Adv Exp Med Biol. 2020;1273:69–90. [DOI] [PubMed] [Google Scholar]
- [50].Gay CM, Stewart CA, Park EM, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell. 2021;39:346–360.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]