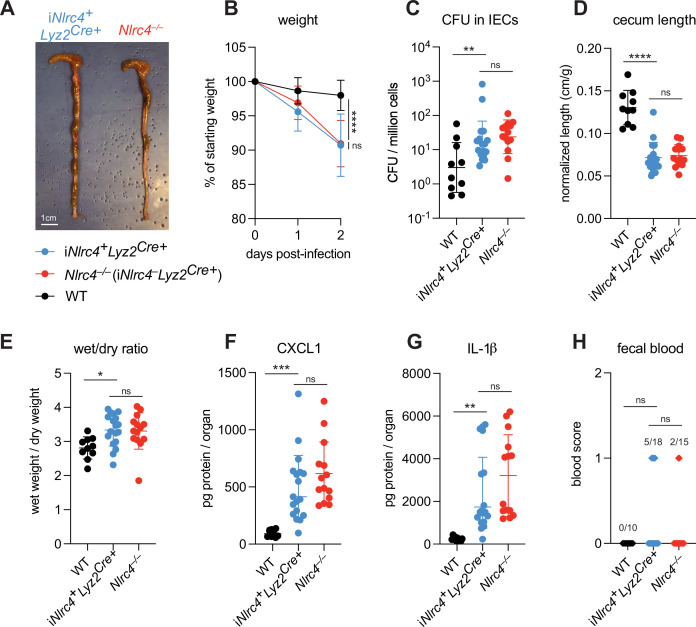
Figure 5. NLRC4 in myeloid-derived cells does not affect Shigella pathogenesis.
(A–H) B6.WT (black, n=10) mice were co-housed with B6.iNlrc4+Lyz2Cre+ (blue, n=18) and B6.Nlrc4–/– (iNlrc4–Lyz2Cre+, red, n=15) littermates, treated orally with 25 mg streptomycin sulfate in water, and orally challenged the next day with 107 colony forming units (CFUs) of wild-type (WT) Shigella flexneri. Mice were sacrificed at 2 days post-infection. (A) Representative images of the cecum and colon from iNlrc4+Lyz2Cre+ and B6.Nlrc4–/– mice. Note the similarity in gross pathology between the two genotypes. (B) Mouse weights from 0 through 2 days post-infection. Each symbol represents the mean for all mice of the indicated group. (C) Shigella CFUs per million cells from the combined intestinal epithelial cell (IEC) enriched fraction of gentamicin-treated cecum and colon tissue. (D) Quantification of cecum lengths normalized to mouse weight prior to infection; cecum length (cm)/mouse weight (g). (E) The ratio of fecal pellet weight when wet (fresh) divided by the fecal pellet weight after overnight drying. Pellets were collected at day 2 post-infection. (F, G) CXCL1 and IL-1β levels measured by ELISA from homogenized cecum and colon tissue of infected mice. (H) Blood scores from feces collected at 2 days post-infection. 1=occult blood, 2=macroscopic blood. (C–H) Each symbol represents one mouse. Data collected from two independent experiments. Mean ± SD is shown in (B, D–G). Geometric mean ± SD is shown in (C). Statistical significance was calculated by one-way ANOVA with Tukey’s multiple comparison test (B (day 2), C–G) and by Fisher’s exact test in (H) where data were stratified by presence (score = 1 or 2) or absence (score = 0) of blood. Data were log-transformed prior to calculations in (C) to achieve normality. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns = not significant (p>0.05).