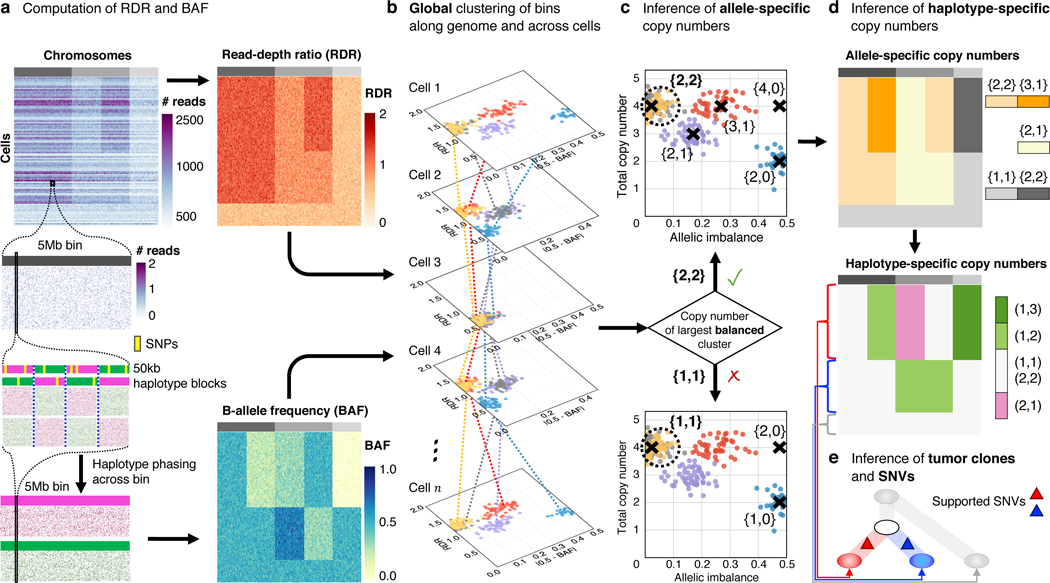
Fig. 1: The CHISEL algorithm.
a, CHISEL computes RDRs and BAFs in low-coverage ( per cell) single-cell DNA sequencing data (top left). Read counts from individual cells (rows) in Mb genomic bins (columns) across three chromosomes (grey rectangles in first row) are shown. For each bin in each cell, CHISEL computes the RDR (top) by normalizing the observed read counts. CHISEL computes the BAF in each bin and cell (bottom) by first performing referenced-based phasing of germline SNPs in kb haplotype blocks (magenta and green) and then phasing all these blocks jointly across all cells. b, CHISEL clusters RDRs and BAFs globally along the genome and jointly across all cells resulting here in 5 clusters of genomic bins (red, blue, purple, yellow, and grey) with distinct copy-number states. c, CHISEL infers a pair of allele-specific copy numbers for each cluster by determining whether the allele-specific copy numbers of the largest balanced (BAF ) cluster are equal to (diploid), (tetraploid), or are higher ploidy. d, CHISEL infers haplotype-specific copy numbers by phasing the allele-specific copy numbers consistently across all cells. e, CHISEL clusters tumor cells into clones according to their haplotype-specific copy numbers. Here, a diploid clone (light gray) and two tumor clones (red and blue) are obtained. A phylogenetic tree describes the evolution of these clones. Somatic SNVs are derived from pseudo-bulk samples and placed on the branches of the tree.