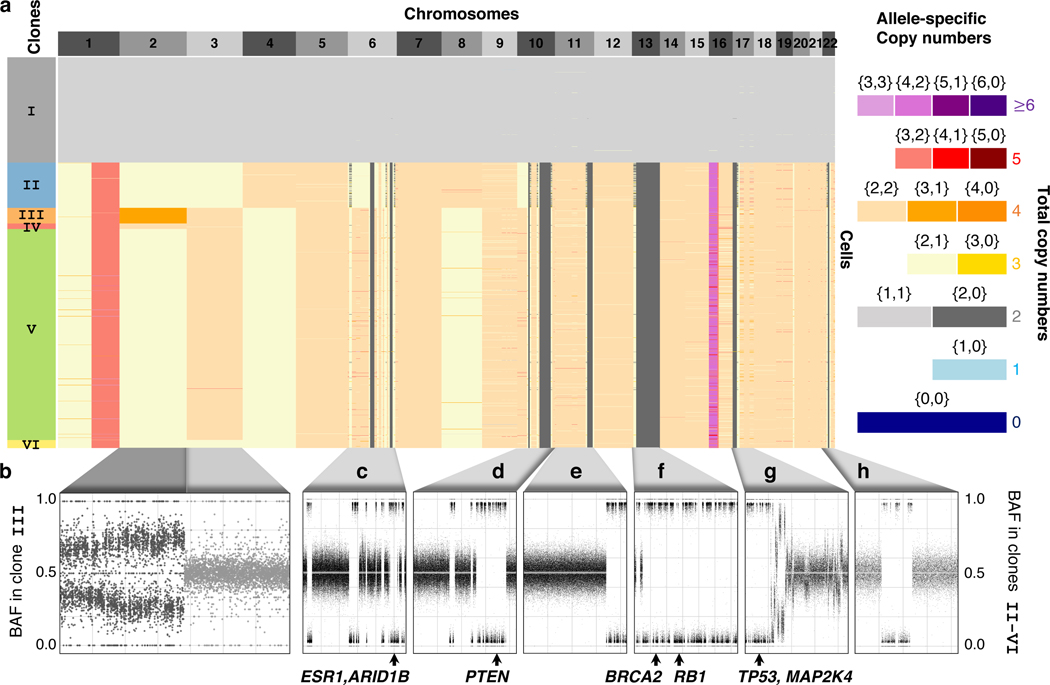
Fig. 2: CHISEL reliably identifies allele-specific copy numbers.
a, Allele-specific copy numbers inferred by CHISEL across 1448 cells from section E of breast cancer patient S0. Clustering of cells according to allele-specific copy numbers reveals 6 clones (colored boxes on left). Clones III and IV are distinguished by an allele-specific CNA affecting the entire chromosome 2; cells in both these clones have 4 total copies of chromosome 2 but distinct allele-specific copy numbers equal to and , respectively. b, The BAF (computed in kb haplotype blocks) of cells from clone III exhibits a clear shift away from BAF=0.5 in chromosome 2, supporting the unequal copy numbers of the two alleles on this chromosome. In contrast, chromosome 3 shows BAF0.5, supporting equal copy numbers of the two alleles. c-h, Copy-neutral LOHs are the most frequent allele-specific CNAs identified by CHISEL with examples shown on six chromosomes. All of these regions have a total copy number equal to 2, the same as the cells in the diploid clone I, but have allele-specific copy numbers equal to . Each of these copy-neutral LOHs is supported by BAFs approximately equal to 0 or 1 indicating the complete loss of one allele across all tumors cells. Known breast-cancer genes in each LOH region are indicated below each plot.