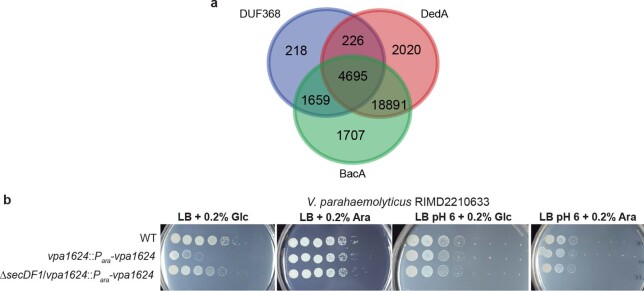
Extended Data Fig. 10. Distribution of putative C55-P translocases in bacteria and genetic analysis of a DUF368 protein in V. parahaemolyticus.
(a): Venn diagram of Annotree species-level outputs for PFAM queries PF04018 (DUF368), PF09335 (DedA), and PF02673 (BacA/UppP). Out of 30,255 bacterial species in the Annotree database, 29,416 (97.2%) are predicted to contain at least one of the three proteins. A list of species in each Venn diagram segment is listed in Supplementary Table 9. This is likely also an overestimate of the number of species lacking any of these three domains due to sequence divergence and potentially inadequate PFAM annotation. For example, the obligate intracellular pathogen Rickettsia rickettsii (GCA_000018225.1) is in the triply-absent group, but upon manual curation is known to have peptidoglycan and has an annotated DedA coding sequence, perhaps reflecting a PFAM annotation lag. Interestingly, however, Mycoplasma species, which do not have lipopolysaccharide or peptidoglycan (and thus may not be dependent on C55-P translocation) appear to be genuinely triply-absent except for a select few members with only a DUF368-containing protein. (b): Growth of V. parahaemolyticus lacking the DUF368-containing protein VPA1624 at neutral and acidic pH. VPA1624 genetically interacts with the V. parahaemolyticus secDF1 loci, as deletion of these suppressors originally identified in V. cholerae rescues the vpa1624-depleted strain. Ara: arabinose, Glc: glucose. Representative data of n = 3 independent cultures per strain/condition.