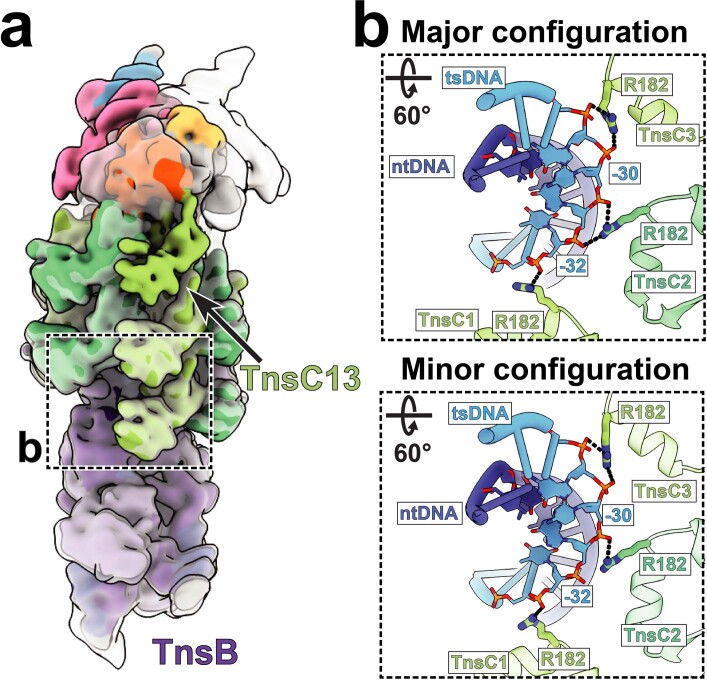
Extended Data Fig. 8. TnsC-DNA interaction proximal to TnsB is identical in the major and minor configurations.
a. Low pass filtered (10 Å) cryo-EM reconstructions of both major (transparent grey) and minor (solid, colored) configurations are aligned with respect to TnsB. The dashed box indicates the TnsC-DNA interactions at the TnsB proximal region shown as inset in panel B. b. Three TnsB-proximal TnsC protomers (From TnsC1 to TnsC3) in both major and minor configurations interact with target strand DNA (tsDNA) in an identical manner through residue R182. Hydrogen bonding interactions (distance cut off <4 Å) between protein residues and the sugar-phosphate backbone of DNA are represented with dashed lines. Base pairs that are interacting with TnsC are represented as filled nucleotides and stick phosphate-backbone.