Fig. 6. Mechanistic model of recruitment and integration of the transposition components in ShCAST.
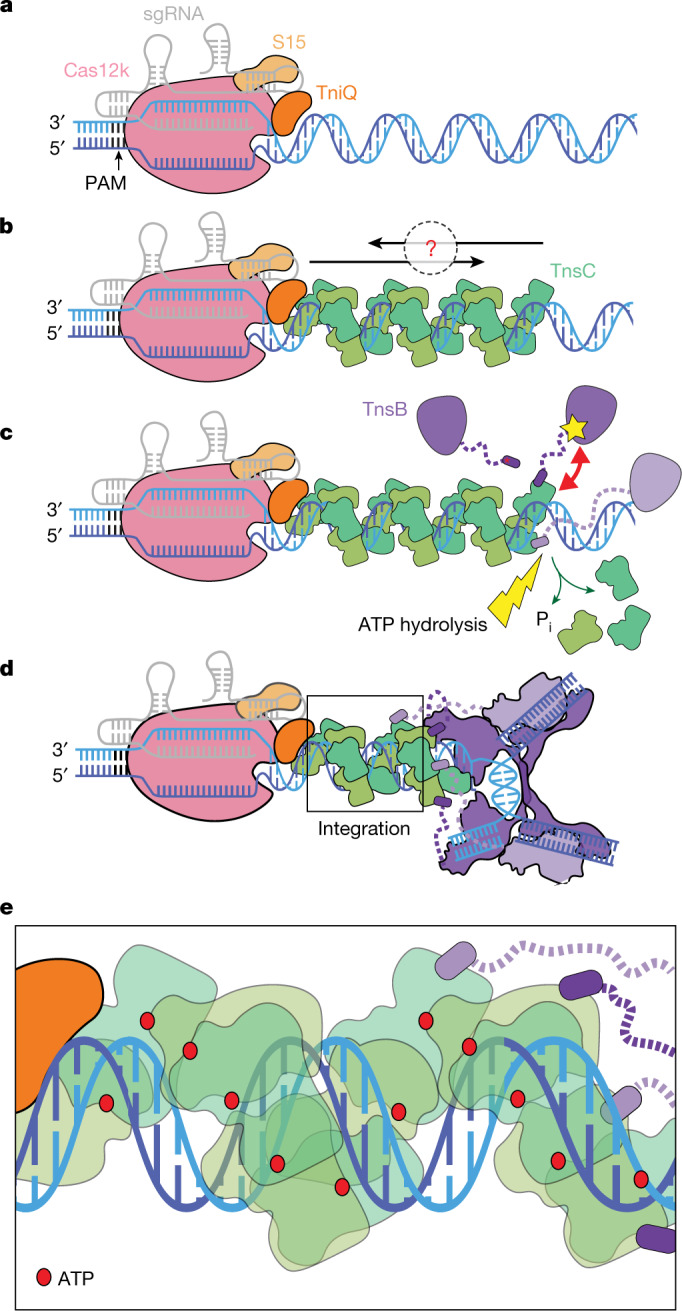
a, The CRISPR effector Cas12k (pink) bound to sgRNA (grey) defines the target site by forming an R-loop and associating with S15 (tan) and TniQ (orange). b, TnsC (green) may polymerize on DNA (blue) in two directions: towards or away from the target site (indicated by the black arrows and the question mark). TnsC interacts with target-site-associated proteins (Cas12k–sgRNA, S15 and TniQ) to form the recruitment complex. c, Two interactions between TnsB (purple) and TnsC: TnsB is recruited to TnsC filaments by TnsBhook (purple, indicated with a red asterisk) and domain IIβ (indicated approximately by a yellow star) from TnsB can interact with TnsC (indicated by a red double arrow), lead to disassembly of TnsC and promote ATP hydrolysis (yellow lightning bolt), resulting in the release of phosphate (green arrows). Dashed lines (purple) represent flexible linkers between TnsBhook and the rest of TnsB. d, TnsB is recruited to the target site and forms the STC upon integration. Four TnsBhook are bound to four TnsC protomers proximal to TnsB through the flexible linker (dashed purple line). Two turns of TnsC (12 protomers) are stably formed against disassembly. Together with target-site-associated proteins and TnsB STC, all CAST components form the transpososome at the integration site. e, A magnified view of TnsC (boxed) in the transpososome shown in d. All TnsC protomers are in the ATP-bound state (ATP is shown as a red circle). TnsC in the transpososome does not specifically track with DNA helical symmetry, unlike previous helical structures of TnsC.
