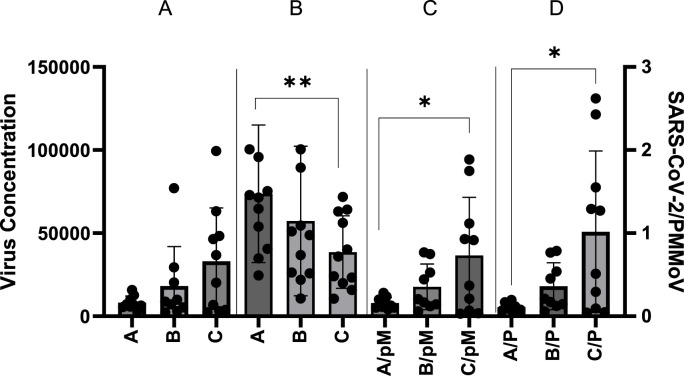
Fig. 1.
SARS-CoV-2 and PMMOV wastewater concentration for communities. Left axis- RNA genome copies concentration. Right axis- SARS-CoV-2 genome copies per liter (L) normalization by dividing by quantified PMMoV RNA per 5 μL. The four sections are A) unadjusted SARS-CoV-2 per L, B) PMMoV per 5 μL, C) SARS-CoV-2 adjusted with PMMoV by accounting for the median PMMoV per 5 μL, and D) SARS-CoV-2 concentration when adjusting for the population by the daily PMMoV concentration measured from communities A, B, and C. Wastewater collection occurred at combined manhole sites that collected wastewater from each community before integration with the main city sewer line. All raw/PMMoV-normalized N1 samples from the collection period were included (represented by the dots) from Communities A, B and C. SARS-CoV-2 was normalized (first normalization) to the ratio of the site PMMoV concentration on collection to the median PMMoV concentration of each site (RNA per PCR reaction) as well as normalized to the daily concentration of PMMoV (second normalization). Significance of measurable difference determined from Kruskal-Wallis chi-square test of means. The mean concentration is represented by the bars. The error bars represent one standard deviation of the mean. The Pairwise comparison determined by Wilcoxon rank sum exact test. *p < 0.05, **p < 0.01, pM = PMMoV median, P=PMMoV RNA genome copies per 5 μL.