FIGURE 2.
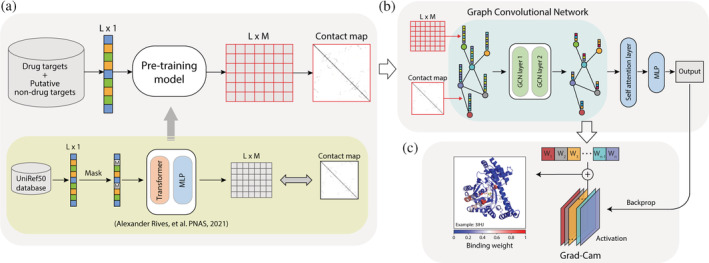
Flowchart for the drug target protein‐prediction algorithm QuoteTarget developed in this study. (a) Protein pretraining model and coding steps. All protein sequences were encoded into a matrix and a contact map as inputs for subsequent classifiers (top panel). This pretraining model was adapted from Rives et al. (bottom panel). (b) Training graph convolutional neural network (GCN) for drug target protein classification. The matrices generated in the coding step were used as the nodes of the graph, and the contact maps were used as the edges of the graph. (c) The drug molecular‐binding weight of each residue was calculated by grad–cam algorithm. Left panel shows an example of protein 3IHJ. The color represents the drug's molecular‐binding weight
