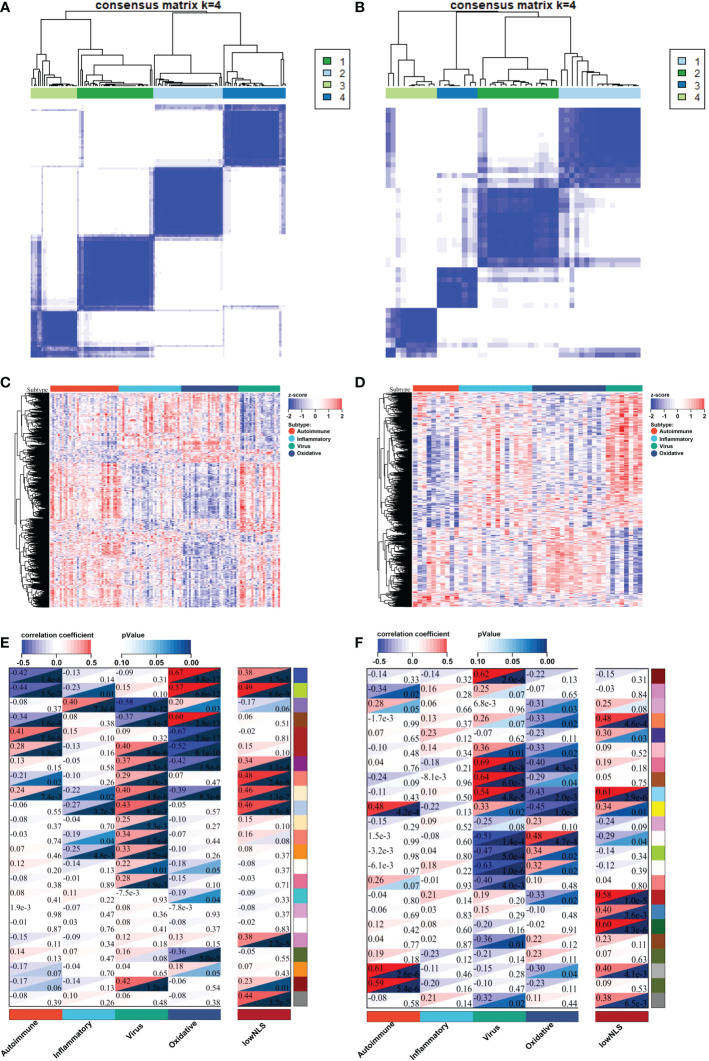
Figure 2.
Identify four molecular subtypes of patients with BA in the training and validation datasets. By applying SNF-CC, training set (A) and validation set (B) were clustered into four distinct subtypes. GSVA similarly classified the samples into four subtypes in training set (C) and validation set (D). Each subtype in training set (E) and validation set (F) was involved with different gene modules of WGCNA. BA, biliary atresia; NLS, natural liver survival; SNF-CC, Similarity Network Fusion and Consensus clustering; GSVA, Gene set variation analysis; WGCNA, Weighted Gene Co-expression Network Analysis.