FIG 4.
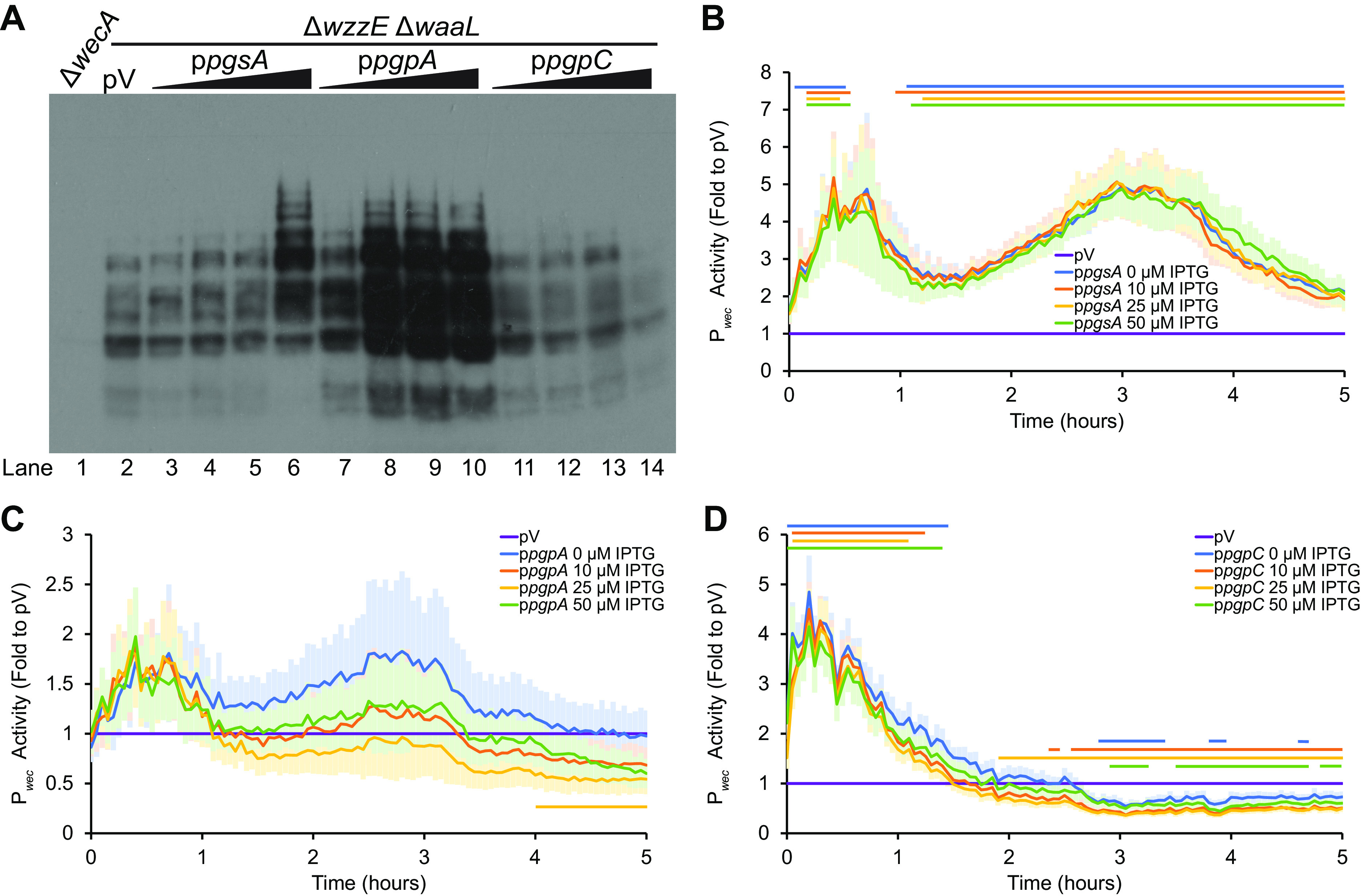
The substrate for ECAPG biosynthesis is in the PG/CL biosynthesis pathway. (A) ECAPG levels were assayed by immunoblot analysis in strains overexpressing the indicated genes in phospholipid biosynthesis. The ΔwzzE ΔwaaL strain background allows the production of ECAPG without the other forms of ECA. For phospholipid synthesis gene overexpression, cultures were treated with 0, 10, 25, and 50 μM IPTG. The vector control (pV) was treated with 50 μM IPTG. A ΔwecA strain serves as a negative control. The image is representative of three independent experiments. Overexpression of both pgsA and pgpA increase accumulation of ECAPG. Overexpression of pgpC causes a small decrease in ECAPG levels. (B to D) Pwec activity was assayed in strains overexpressing genes in phospholipid biosynthesis as in Fig. 3. The means ± the SEM for the relative luminescence as a fold value to the vector control are shown for six biological replicates. pV, empty vector sample carrying the Pwec reporter and the empty vector for the phospholipid synthesis gene overexpression and treated with 50 μM IPTG. Horizontal bars: P < 0.05 by t test consistently for three or more time points. (B) Overexpressing pgsA consistently increased Pwec activity. (C) Overexpression of pgpA did not change Pwec activity at most time points and IPTG concentrations. (D) Overexpressing pgpC causes early increases and later decreases in Pwec activity.
